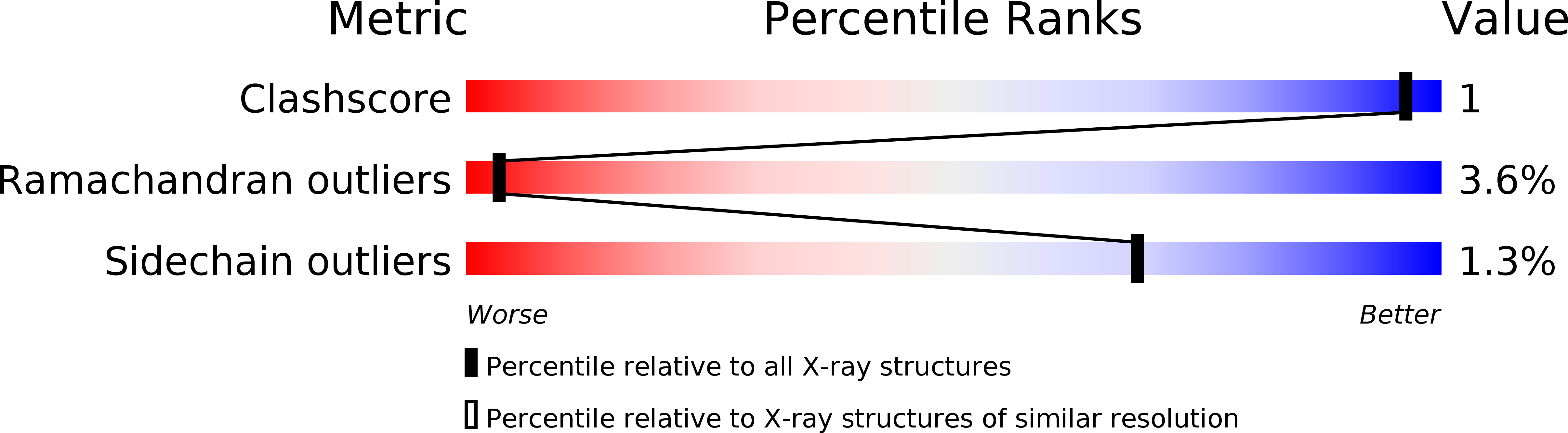Conformational mapping of the N-terminal peptide of HIV-1 gp41 in lipid detergent and aqueous environments using 13C-enhanced Fourier transform infrared spectroscopy.
Gordon, L.M., Mobley, P.W., Lee, W., Eskandari, S., Kaznessis, Y.N., Sherman, M.A., Waring, A.J.(2004) Protein Sci 13: 1012-1030
- PubMed: 15044732
- DOI: https://doi.org/10.1110/ps.03407704
- Primary Citation of Related Structures:
1P5A - PubMed Abstract:
The N-terminal domain of HIV-1 glycoprotein 41,000 (gp41) participates in viral fusion processes. Here, we use physical and computational methodologies to examine the secondary structure of a peptide based on the N terminus (FP; residues 1-23) in aqueous and detergent environments. (12)C-Fourier transform infrared (FTIR) spectroscopy indicated greater alpha-helix for FP in lipid-detergent sodium dodecyl sulfate (SDS) and aqueous phosphate-buffered saline (PBS) than in only PBS. (12)C-FTIR spectra also showed disordered FP conformations in these two environments, along with substantial beta-structure for FP alone in PBS. In experiments that map conformations to specific residues, isotope-enhanced FTIR spectroscopy was performed using FP peptides labeled with (13)C-carbonyl. (13)C-FTIR results on FP in SDS at low peptide loading indicated alpha-helix (residues 5 to 16) and disordered conformations (residues 1-4). Because earlier (13)C-FTIR analysis of FP in lipid bilayers demonstrated alpha-helix for residues 1-16 at low peptide loading, the FP structure in SDS micelles only approximates that found for FP with membranes. Molecular dynamics simulations of FP in an explicit SDS micelle indicate that the fraying of the first three to four residues may be due to the FP helix moving to one end of the micelle. In PBS alone, however, electron microscopy of FP showed large fibrils, while (13)C-FTIR spectra demonstrated antiparallel beta-sheet for FP (residues 1-12), analogous to that reported for amyloid peptides. Because FP and amyloid peptides each exhibit plaque formation, alpha-helix to beta-sheet interconversion, and membrane fusion activity, amyloid and N-terminal gp41 peptides may belong to the same superfamily of proteins.
Organizational Affiliation:
REI at Harbor-UCLA Medical Center, 124 West Carson Street, Bldg. F5 South, Torrance, CA 90502-2064, USA. lgordon2@san.rr.com