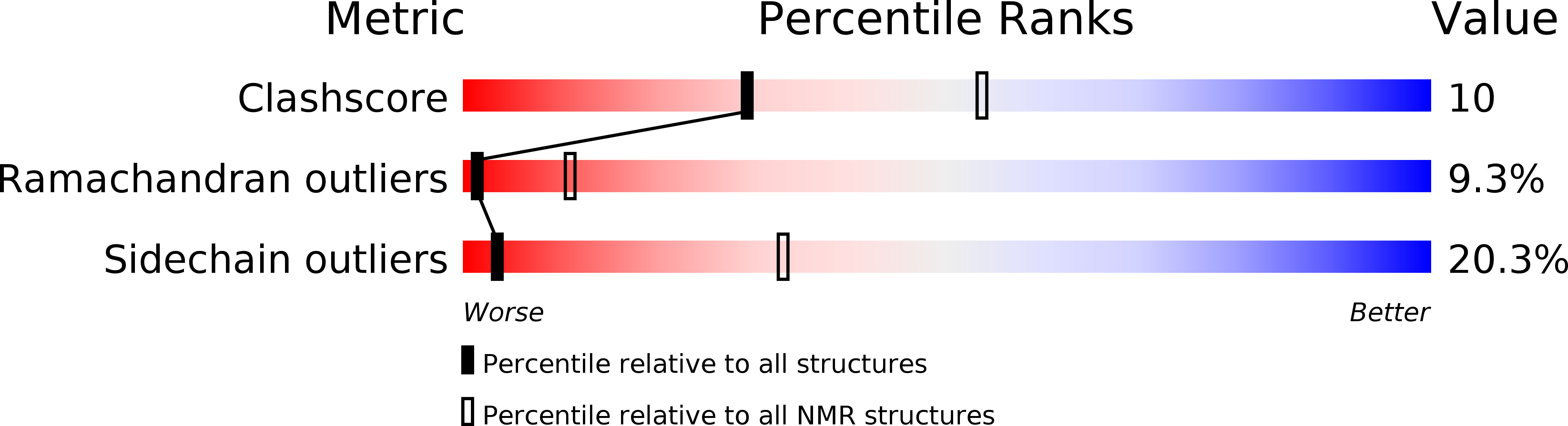The solution structure of gomesin, an antimicrobial cysteine-rich peptide from the spider.
Mandard, N., Bulet, P., Caille, A., Daffre, S., Vovelle, F.(2002) Eur J Biochem 269: 1190-1198
- PubMed: 11856345
- DOI: https://doi.org/10.1046/j.0014-2956.2002.02760.x
- Primary Citation of Related Structures:
1KFP - PubMed Abstract:
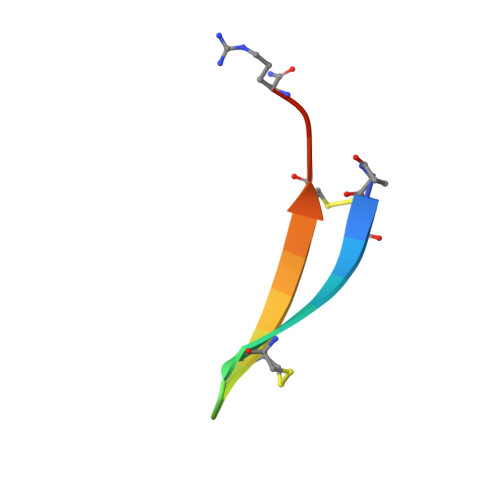
Gomesin is the first peptide isolated from spider exhibiting antimicrobial activities. This highly cationic peptide is composed of 18 amino-acid residues including four cysteines forming two disulfide linkages. The solution structure of gomesin has been determined using proton two-dimensional NMR (2D-NMR) and restrained molecular dynamics calculations. The global fold of gomesin consists in a well-resolved two-stranded antiparallel betasheet connected by a noncanonical betaturn. A comparison between the structures of gomesin and protegrin-1 from porcine and androctonin from scorpion outlines several common features in the distribution of hydrophobic and hydrophilic residues. The N- and C-termini, the betaturn and one face of the betasheet are hydrophilic, but the hydrophobicity of the other face depends on the peptide. The similarities suggest that the molecules interact with membranes in an analogous manner. The importance of the intramolecular disulfide bridges in the biological activity of gomesin is being investigated.
Organizational Affiliation:
Centre de Biophysique Moléculaire, CNRS, Orléans, France.