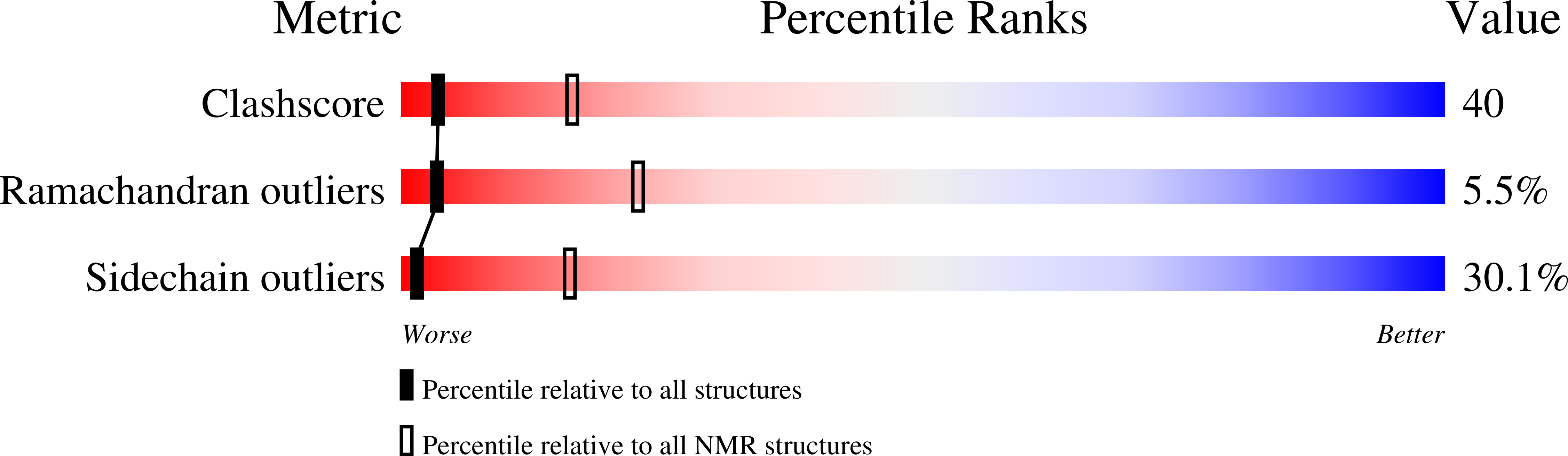Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
Hemmi, H., Kumazaki, T., Yamazaki, T., Kojima, S., Yoshida, T., Kyogoku, Y., Katsu, M., Shinohara, F., Yokosawa, H., Miura, K., Kobayashi, Y.(2003) Biochemistry 42: 2524-2534
- PubMed: 12614146
- DOI: https://doi.org/10.1021/bi026727c
- Primary Citation of Related Structures:
1IY5, 1IY6 - PubMed Abstract:
The ovomucoid third domain from silver pheasant (OMSVP3), a typical Kazal-type inhibitor, strongly inhibits different serine proteases of various specificities, i.e., chymotrypsin, Streptomyces griseus protease, subtilisin, and elastase. Structural studies have suggested that conformational flexibility in the reactive site loop of the free inhibitor may be related to broad specificity of the ovomucoid. On the basis of the structural homology between OMSVP3 and ascidian trypsin inhibitor (ATI), which has a cystine-stabilized alpha-helical (CSH) motif in the sequence, we prepared the disulfide variant of OMSVP3, introducing an engineered disulfide bond between positions 14 and 39 near the reactive site (Met18-Glu19) by site-directed mutagenesis. The disulfide variant P14C/N39C retained potent inhibitory activities toward alpha-chymotrypsin (CHT) and S. griseus proteases A and B (SGPA and SGPB), while this variant lost most of its inhibitory activity toward porcine pancreatic elastase (PPE). We determined the solution structure of P14C/N39C, as well as that of wild-type OMSVP3, by two-dimensional nuclear magnetic resonance (2D NMR) methods and compared their structures to elucidate the structural basis of the inhibitory specificity change. For the molecular core consisting of a central alpha-helix and a three-stranded antiparallel beta-sheet, essentially no structural difference was detected between the two (pairwise rmsd value = 0.41 A). In contrast to this, a significant difference was detected in the loop from Cys8 to Thr17, where in P14C/N39C it has drawn approximately 4 A nearer the central helix to form the engineered Cys14-Cys39 bond. Concomitantly, the Tyr11-Pro12 cis-peptide linkage, which is highly conserved in ovomucoid third domains, was isomerized to the trans configuration. Such structural change in the loop near the reactive site may possibly affect the inhibitory specificity of P14C/N39C for the corresponding proteases.
Organizational Affiliation:
National Food Research Institute, 2-1-12 Kannondai, Tsukuba, Ibaraki 305-8642, Japan.