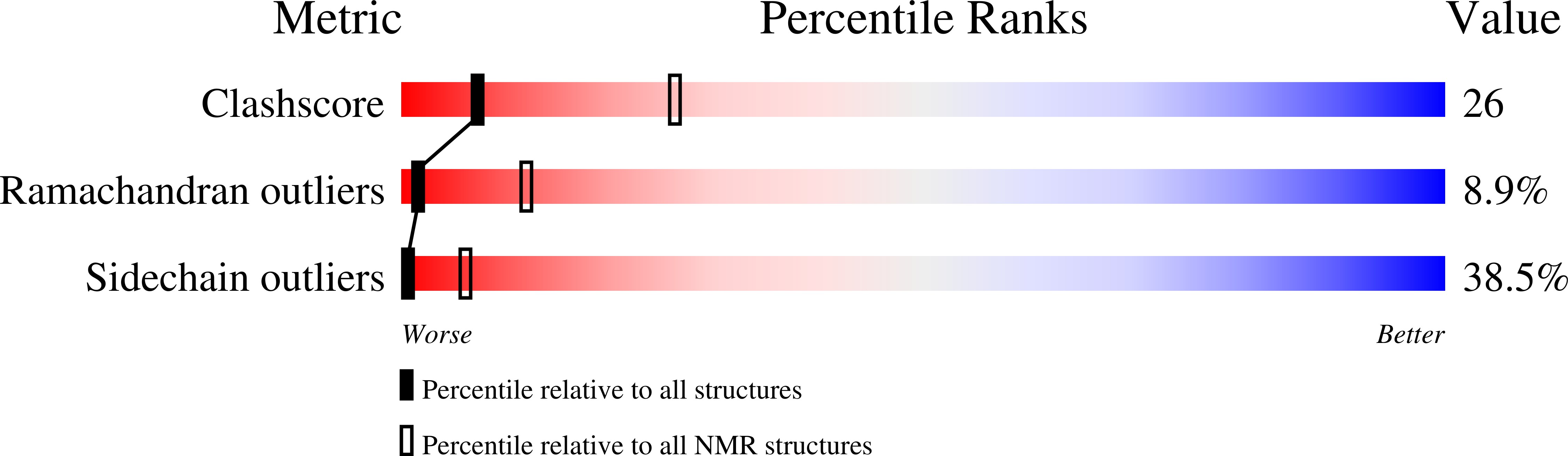Solution structure of Ptu1, a toxin from the assassin bug Peirates turpis that blocks the voltage-sensitive calcium channel N-type.
Bernard, C., Corzo, G., Mosbah, A., Nakajima, T., Darbon, H.(2001) Biochemistry 40: 12795-12800
- PubMed: 11669615
- DOI: https://doi.org/10.1021/bi015537j
- Primary Citation of Related Structures:
1I26 - PubMed Abstract:
Ptu1 is a toxin from the assassin bug Peirates turpis which has been demonstrated to bind reversibly the N-type calcium channels and to have lower affinity than the omega-conotoxin MVIIA. We have determined the solution structure of Ptu1 by use of conventional two-dimensional NMR techniques followed by distance-geometry and molecular dynamics. The calculated structure of Ptu1 belongs to the inhibitory cystin knot structural family (ICK) that consists of a compact disulfide-bonded core from which four loops emerge. Analysis of the 25 converged solutions indicates that the molecular structure of Ptu1 contains a 2-stranded antiparallel beta-sheet (residues 24-27 and 31-34) as the only secondary structure. The loop 2 that has been described to be critical for the binding of the toxin on the channel is similar in Ptu1 and MVIIA. In this loop, the critical residue, Tyr13, in MVIIA is retrieved in Ptu1 as Phe13, but the presence of an acidic residue (Asp16) in Ptu1 could disturb the binding of Ptu1 on the channel and could explain the lower affinity of Ptu1 toward the N-type calcium channel compared to the one of MVIIA. Analysis of the electrostatic charge's repartition gives some insights about the importance of the basic residues, which could interact with acidic residues of the channel and then provide a stabilization of the toxin on the channel.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques, UMR 6098, CNRS and Universités d'Aix-Marseille I and II, 31 Chemin Joseph Aiguier, 13402 Marseille Cedex 20, France.