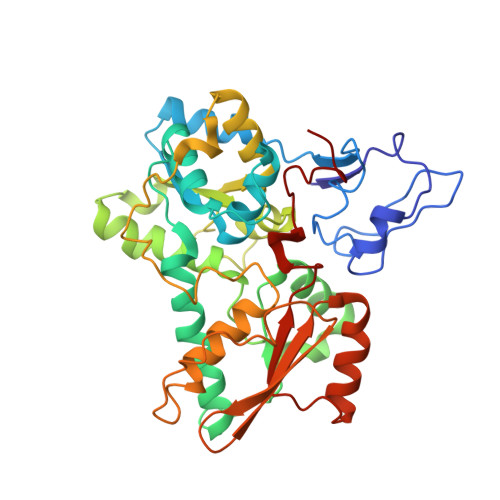
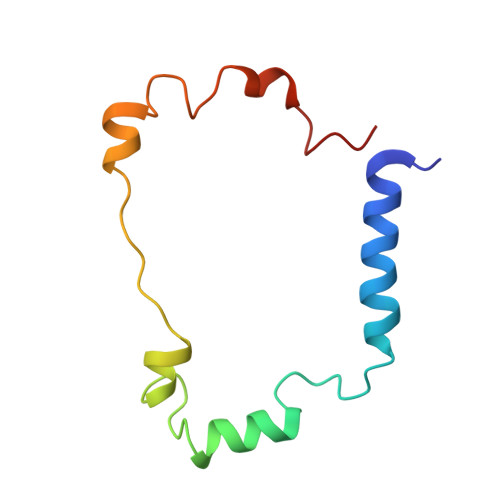
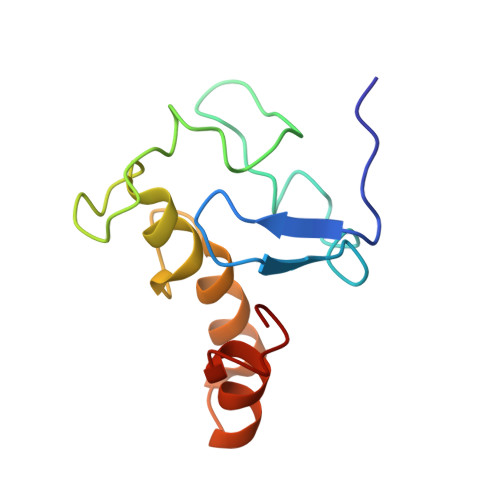
The Cytochrome C(3)-[Fe]-Hydrogenase Electron-Transfer Complex: Structural Model by NMR Restrained Docking
Elantak, L., Morelli, X., Bornet, O., Hatchikian, C., Czjzek, M., Dolla, A., Guerlesquin, F.(2003) FEBS Lett 548: 1
- PubMed: 12885397
- DOI: https://doi.org/10.1016/s0014-5793(03)00718-x
- Primary Citation of Related Structures:
1GX7 - PubMed Abstract:
Cytochrome c(3) (M(r) 13000) is a low redox potential cytochrome specific of the anaerobic metabolism in sulfate-reducing bacteria. This tetrahemic cytochrome is an intermediate between the [Fe]-hydrogenase and the cytochrome Hmc in Desulfovibrio vulgaris Hildenborough strain. The present work describes the structural model of the cytochrome c(3)-[Fe]-hydrogenase complex obtained by nuclear magnetic resonance restrained docking. This model connects the distal cluster of the [Fe]-hydrogenase to heme 4 of the cytochrome, the same heme found in the interaction with cytochrome Hmc. This result gives evidence that cytochrome c(3) is an electron shuttle between the periplasmic hydrogenase and the Hmc membrane-bound complex.
Organizational Affiliation:
Unité de Bioénergétique et Ingénierie des Protéines, IBSM-CNRS, 31 chemin Joseph Aiguier, 13402 Cedex 20, Marseille, France. guerlesq@ibsm.cnrs-mrs.fr