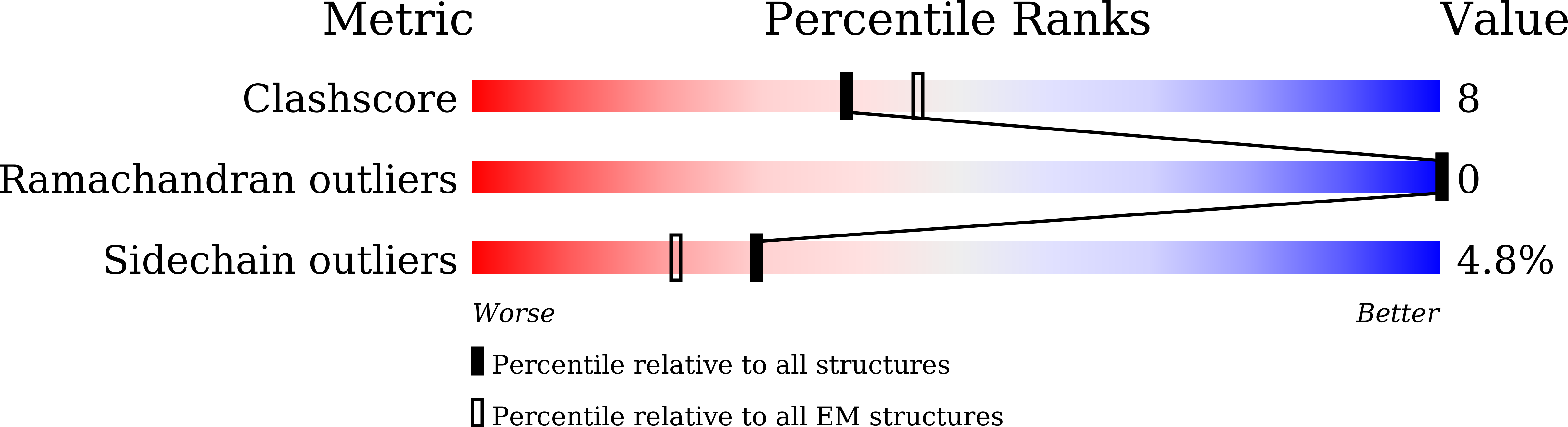Molecular basis of hippopotamus ACE2 binding to SARS-CoV-2.
Yang, R., Han, P., Han, P., Li, D., Zhao, R., Niu, S., Liu, K., Li, S., Tian, W.-.X., Gao, G.F.(2024) J Virol 98: e0045124-e0045124
- PubMed: 38591877
- DOI: https://doi.org/10.1128/jvi.00451-24
- Primary Citation of Related Structures:
8WLO, 8WLR - PubMed Abstract:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has a wide range of hosts, including hippopotami, which are semi-aquatic mammals and phylogenetically closely related to Cetacea. In this study, we characterized the binding properties of hippopotamus angiotensin-converting enzyme 2 (hiACE2) to the spike (S) protein receptor binding domains (RBDs) of the SARS-CoV-2 prototype (PT) and variants of concern (VOCs). Furthermore, the cryo-electron microscopy (cryo-EM) structure of the SARS-CoV-2 PT S protein complexed with hiACE2 was resolved. Structural and mutational analyses revealed that L30 and F83, which are specific to hiACE2, played a crucial role in the hiACE2/SARS-CoV-2 RBD interaction. In addition, comparative and structural analysis of ACE2 orthologs suggested that the cetaceans may have the potential to be infected by SARS-CoV-2. These results provide crucial molecular insights into the susceptibility of hippopotami to SARS-CoV-2 and suggest the potential risk of SARS-CoV-2 VOCs spillover and the necessity for surveillance. The hippopotami are the first semi-aquatic artiodactyl mammals wherein SARS-CoV-2 infection has been reported. Exploration of the invasion mechanism of SARS-CoV-2 will provide important information for the surveillance of SARS-CoV-2 in hippopotami, as well as other semi-aquatic mammals and cetaceans. Here, we found that hippopotamus ACE2 (hiACE2) could efficiently bind to the RBDs of the SARS-CoV-2 prototype (PT) and variants of concern (VOCs) and facilitate the transduction of SARS-CoV-2 PT and VOCs pseudoviruses into hiACE2-expressing cells. The cryo-EM structure of the SARS-CoV-2 PT S protein complexed with hiACE2 elucidated a few critical residues in the RBD/hiACE2 interface, especially L30 and F83 of hiACE2 which are unique to hiACE2 and contributed to the decreased binding affinity to PT RBD compared to human ACE2. Our work provides insight into cross-species transmission and highlights the necessity for monitoring host jumps and spillover events on SARS-CoV-2 in semi-aquatic/aquatic mammals.
Organizational Affiliation:
College of Veterinary Medicine, Shanxi Agricultural University, Jinzhong, China.