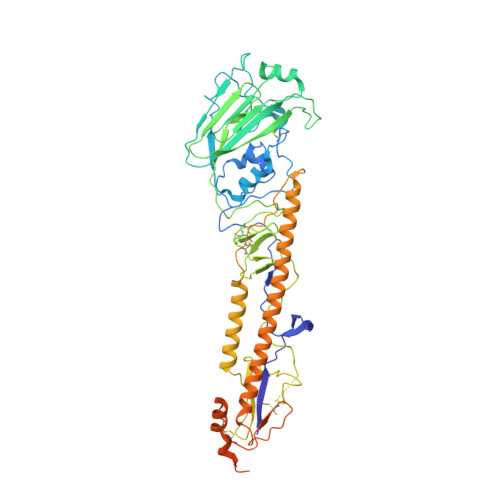
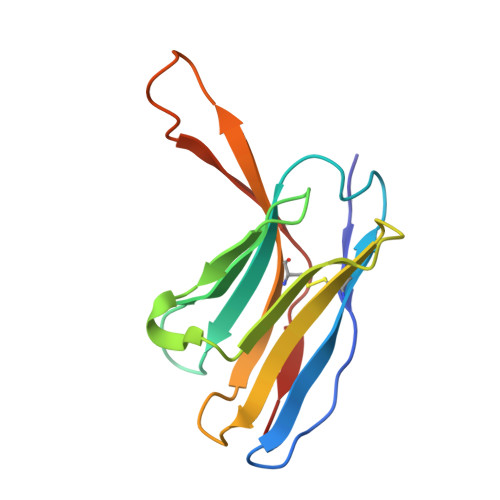
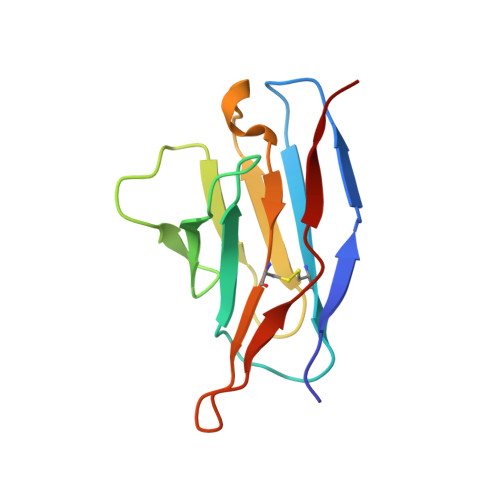
Structural basis for the broad antigenicity of the computationally optimized influenza hemagglutinin X6.
Nagashima, K.A., Dzimianski, J.V., Yang, M., Abendroth, J., Sautto, G.A., Ross, T.M., DuBois, R.M., Edwards, T.E., Mousa, J.J.(2024) Structure
- PubMed: 38810648
- DOI: https://doi.org/10.1016/j.str.2024.05.001
- Primary Citation of Related Structures:
8SJ9, 8V7O - PubMed Abstract:
Influenza causes significant morbidity and mortality. As an alternative approach to current seasonal vaccines, the computationally optimized broadly reactive antigen (COBRA) platform has been previously applied to hemagglutinin (HA). This approach integrates wild-type HA sequences into a single immunogen to expand the breadth of accessible antibody epitopes. Adding to previous studies of H1, H3, and H5 COBRA HAs, we define the structural features of another H1 subtype COBRA, X6, that incorporates HA sequences from before and after the 2009 H1N1 influenza pandemic. We determined structures of this antigen alone and in complex with COBRA-specific as well as broadly reactive and functional antibodies, analyzing its antigenicity. We found that X6 possesses features representing both historic and recent H1 HA strains, enabling binding to both head- and stem-reactive antibodies. Overall, these data confirm the integrity of broadly reactive antibody epitopes of X6 and contribute to design efforts for a next-generation vaccine.
Organizational Affiliation:
Center for Vaccines and Immunology, College of Veterinary Medicine, University of Georgia, Athens, GA, USA; Department of Infectious Diseases, College of Veterinary Medicine, University of Georgia, Athens, GA, USA.