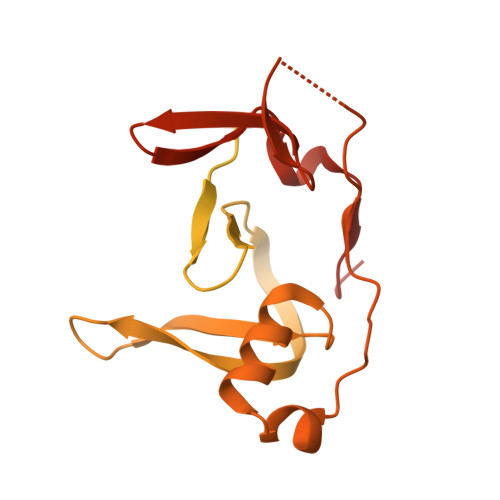
HSV-1 ICP0 dimer domain adopts a novel beta-barrel fold.
McCloskey, E., Kashipathy, M., Cooper, A., Gao, P., Johnson, D.K., Battaile, K.P., Lovell, S., Davido, D.J.(2024) Proteins 92: 830-841
- PubMed: 38372168
- DOI: https://doi.org/10.1002/prot.26673
- Primary Citation of Related Structures:
8UA2, 8UA5 - PubMed Abstract:
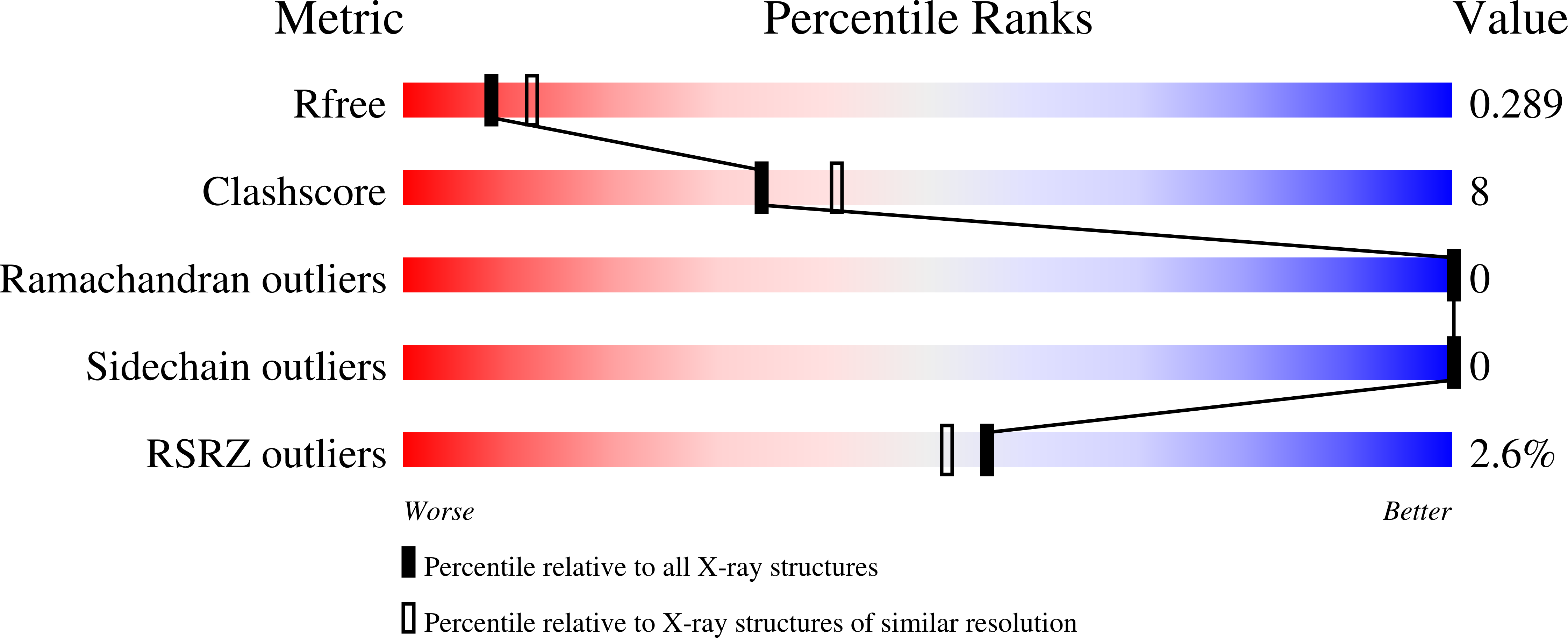
Infected cell protein 0 (ICP0) is an immediate-early regulatory protein of herpes simplex virus 1 (HSV-1) that possesses E3 ubiquitin ligase activity. ICP0 transactivates viral genes, in part, through its C-terminal dimer domain (residues 555-767). Deletion of this dimer domain results in reduced viral gene expression, lytic infection, and reactivation from latency. Since ICP0's dimer domain is associated with its transactivation activity and efficient viral replication, we wanted to determine the structure of this specific domain. The C-terminus of ICP0 was purified from bacteria and analyzed by X-ray crystallography to solve its structure. Each subunit or monomer in the ICP0 dimer is composed of nine β-strands and two α-helices. Interestingly, two adjacent β-strands from one monomer "reach" into the adjacent subunit during dimer formation, generating two β-barrel-like structures. Additionally, crystallographic analyses indicate a tetramer structure is formed from two β-strands of each dimer, creating a "stacking" of the β-barrels. The structural protein database searches indicate the fold or structure adopted by the ICP0 dimer is novel. The dimer is held together by an extensive network of hydrogen bonds. Computational analyses reveal that ICP0 can either form a dimer or bind to SUMO1 via its C-terminal SUMO-interacting motifs but not both. Understanding the structure of the dimer domain will provide insights into the activities of ICP0 and, ultimately, the HSV-1 life cycle.
Organizational Affiliation:
Department of Molecular Biosciences, University of Kansas, Lawrence, Kansas, USA.