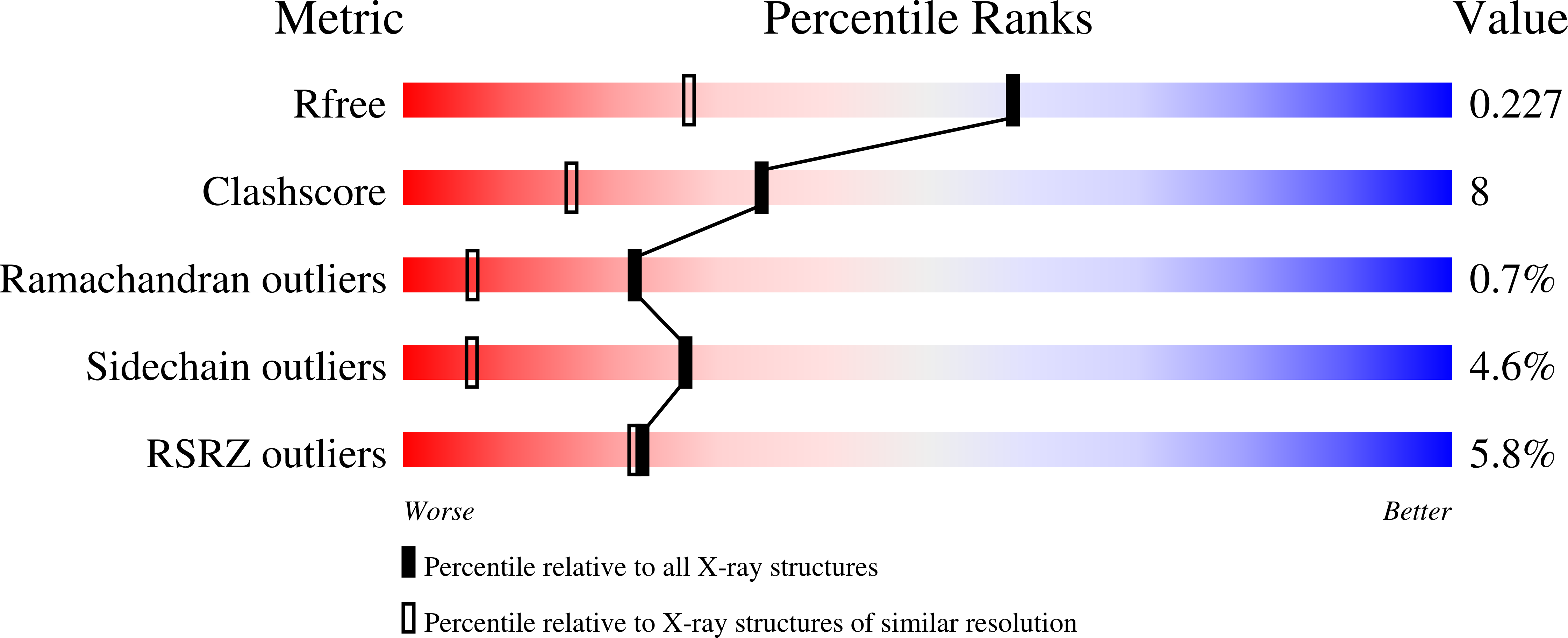
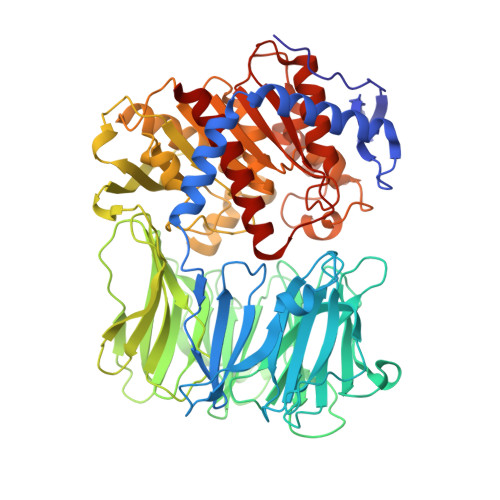
Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution
Petrenko, D.E., Boyko, K.M., Nikolaeva, A.Y., Vlaskina, A.V., Mikhailova, A.G., Timofeev, V.I., Rakitina, T.V.To be published.