Improvement of thermostability and activity of PET-degrading enzyme Cut190 towards a detailed understanding and application of the enzymatic reaction mechanism.
Numoto, N., Kamiya, N., Oda, M.(2023) Biorxiv
Experimental Data Snapshot
(2023) Biorxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Alpha/beta hydrolase family protein | 263 | Saccharomonospora viridis | Mutation(s): 8 Gene Names: Cut190, SAMN02982918_2340 EC: 3.1.1.74 | 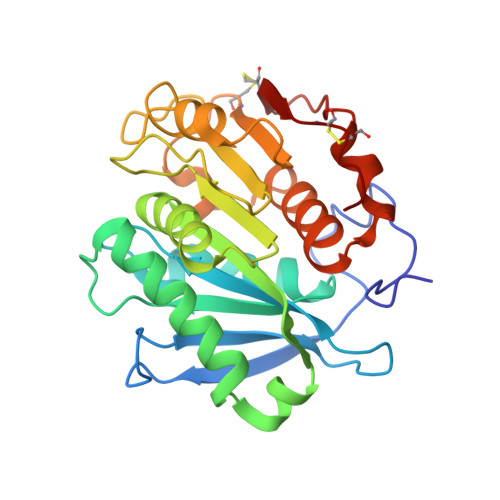 | |
UniProt | |||||
Find proteins for W0TJ64 (Saccharomonospora viridis) Explore W0TJ64 Go to UniProtKB: W0TJ64 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | W0TJ64 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MES (Subject of Investigation/LOI) Query on MES | D [auth A], G [auth B] | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID C6 H13 N O4 S SXGZJKUKBWWHRA-UHFFFAOYSA-N |  | ||
| CA Query on CA | C [auth A], F [auth B] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| CL Query on CL | E [auth A], H [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 83.869 | α = 90 |
| b = 83.869 | β = 90 |
| c = 64.723 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science (JSPS) | Japan | -- |