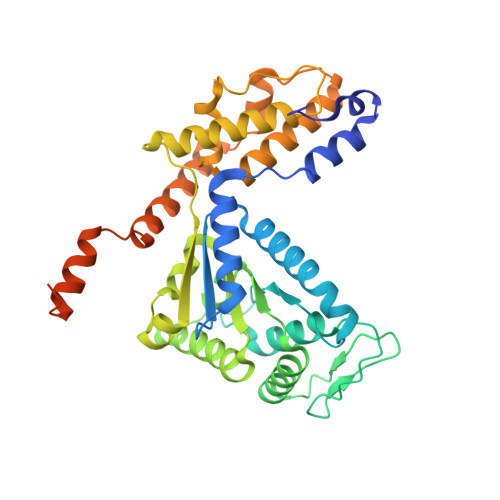
Molecular mechanism for Tn7-like transposon recruitment by a type I-B CRISPR effector.
Wang, S., Gabel, C., Siddique, R., Klose, T., Chang, L.(2023) Cell 186: 4204-4215.e19
- PubMed: 37557170
- DOI: https://doi.org/10.1016/j.cell.2023.07.010
- Primary Citation of Related Structures:
8FCJ, 8FCU, 8FCV, 8FCW, 8FCX, 8FD2, 8FD3, 8FF4, 8FF5 - PubMed Abstract:
Tn7-like transposons have co-opted CRISPR-Cas systems to facilitate the movement of their own DNA. These CRISPR-associated transposons (CASTs) are promising tools for programmable gene knockin. A key feature of CASTs is their ability to recruit Tn7-like transposons to nuclease-deficient CRISPR effectors. However, how Tn7-like transposons are recruited by diverse CRISPR effectors remains poorly understood. Here, we present the cryo-EM structure of a recruitment complex comprising the Cascade complex, TniQ, TnsC, and the target DNA in the type I-B CAST from Peltigera membranacea cyanobiont 210A. Target DNA recognition by Cascade induces conformational changes in Cas6 and primes TniQ recruitment through its C-terminal domain. The N-terminal domain of TniQ is bound to the seam region of the TnsC spiral heptamer. Our findings provide insights into the diverse mechanisms for the recruitment of Tn7-like transposons to CRISPR effectors and will aid in the development of CASTs as gene knockin tools.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, IN 47907, USA.