Predictions of the substrate specificities of the plastic-degrading enzymes IsPETase and AdCut
Clark, M., Zahn, M.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| PBS(A) depolymerase | 271 | Acidovorax delafieldii | Mutation(s): 0 Gene Names: pbsA | 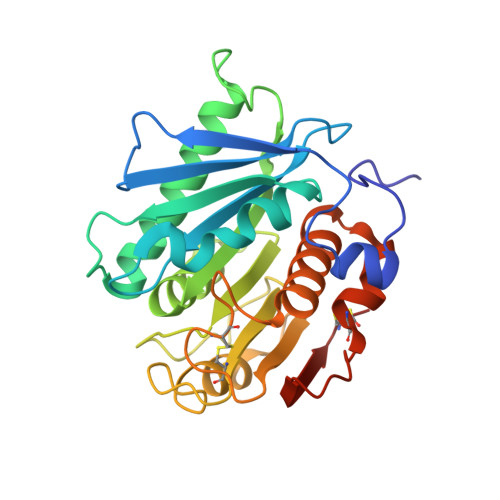 | |
UniProt | |||||
Find proteins for Q8RR62 (Acidovorax delafieldii) Explore Q8RR62 Go to UniProtKB: Q8RR62 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8RR62 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 45.798 | α = 90 |
| b = 96.43 | β = 111.033 |
| c = 58.525 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| autoPROC | data reduction |
| STARANISO | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| UK Research and Innovation (UKRI) | United Kingdom | Research England E3 |