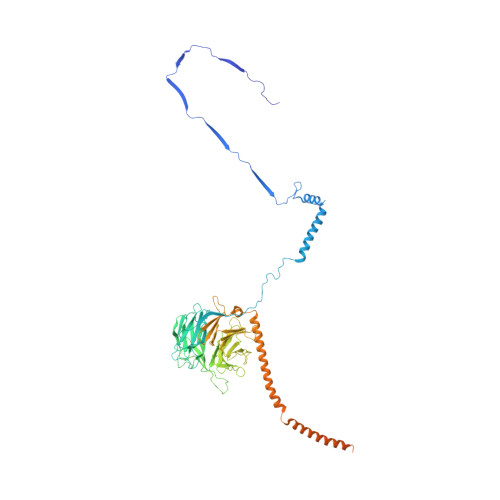
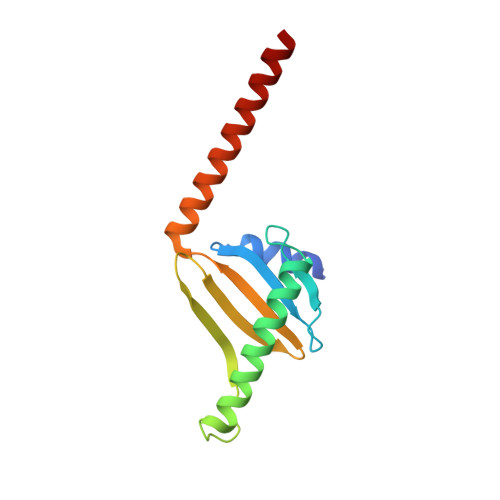
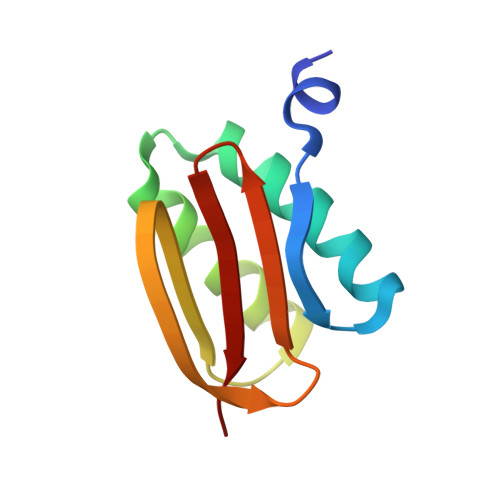
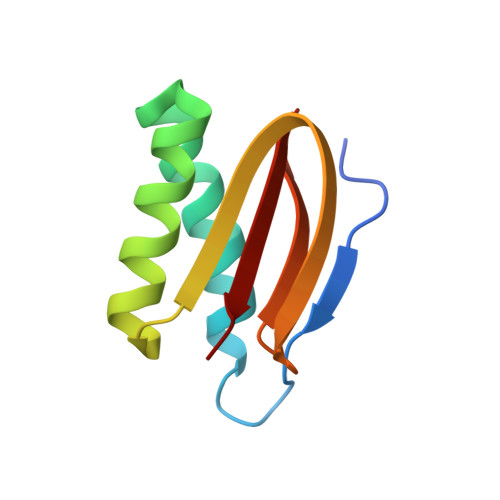
ATP-induced conformational change of axonemal outer dynein arms revealed by cryo-electron tomography.
Zimmermann, N., Noga, A., Obbineni, J.M., Ishikawa, T.(2023) EMBO J 42: e112466-e112466
- PubMed: 37051721
- DOI: https://doi.org/10.15252/embj.2022112466
- Primary Citation of Related Structures:
8BWY, 8BX8 - PubMed Abstract:
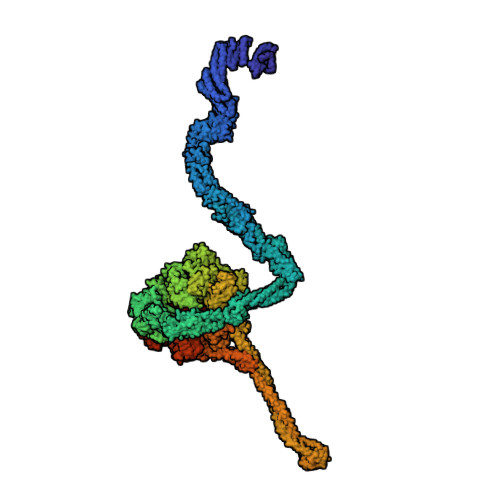
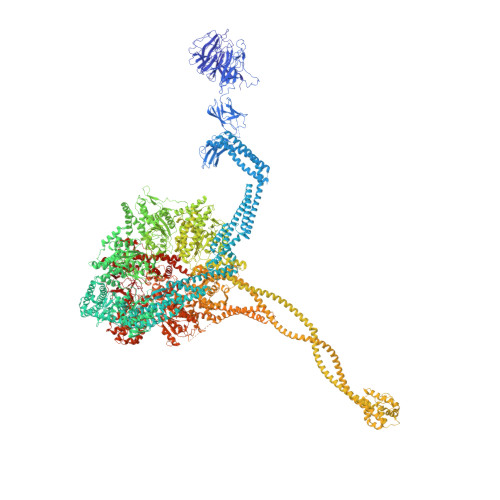
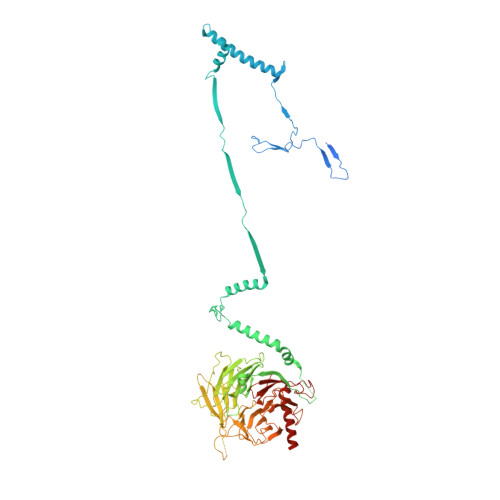
Axonemal outer dynein arm (ODA) motors generate force for ciliary beating. We analyzed three states of the ODA during the power stroke cycle using in situ cryo-electron tomography, subtomogram averaging, and classification. These states of force generation depict the prepower stroke, postpower stroke, and intermediate state conformations. Comparison of these conformations to published in vitro atomic structures of cytoplasmic dynein, ODA, and the Shulin-ODA complex revealed differences in the orientation and position of the dynein head. Our analysis shows that in the absence of ATP, all dynein linkers interact with the AAA3/AAA4 domains, indicating that interactions with the adjacent microtubule doublet B-tubule direct dynein orientation. For the prepower stroke conformation, there were changes in the tail that is anchored on the A-tubule. We built models starting with available high-resolution structures to generate a best-fitting model structure for the in situ pre- and postpower stroke ODA conformations, thereby showing that ODA in a complex with Shulin adopts a similar conformation as the active prepower stroke ODA in the axoneme.
Organizational Affiliation:
Paul Scherrer Institut (PSI), Laboratory of Nanoscale Biology, Villigen PSI, Switzerland.