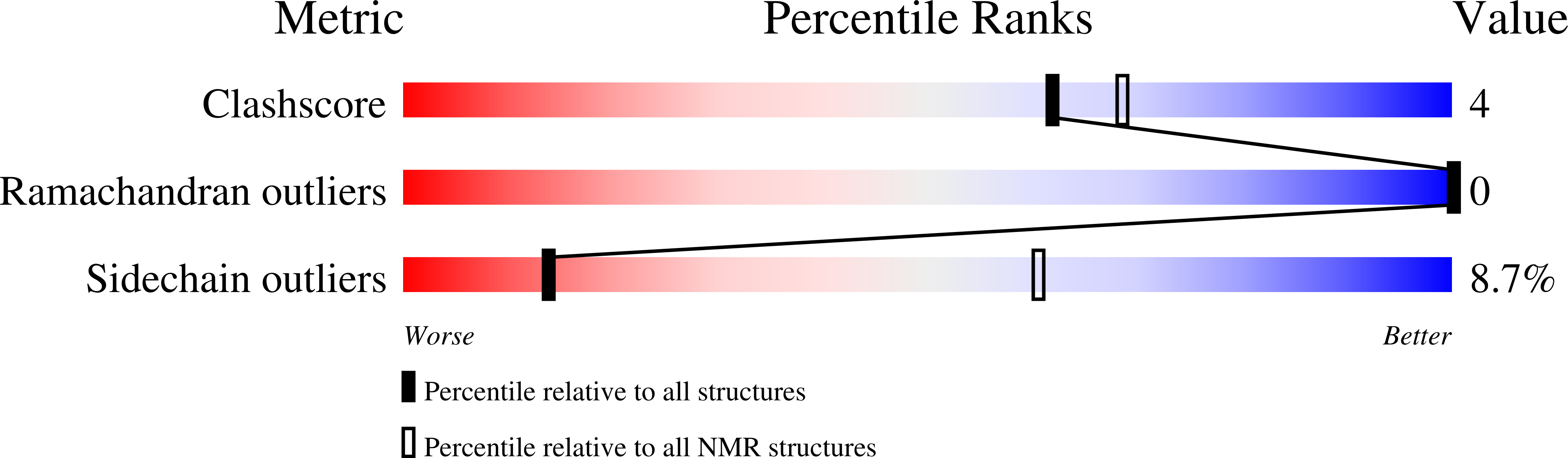A peptide toxin in ant venom mimics vertebrate EGF-like hormones to cause long-lasting hypersensitivity in mammals.
Eagles, D.A., Saez, N.J., Krishnarjuna, B., Bradford, J.J., Chin, Y.K., Starobova, H., Mueller, A., Reichelt, M.E., Undheim, E.A.B., Norton, R.S., Thomas, W.G., Vetter, I., King, G.F., Robinson, S.D.(2022) Proc Natl Acad Sci U S A 119
- PubMed: 35131940
- DOI: https://doi.org/10.1073/pnas.2112630119
- Primary Citation of Related Structures:
7R6P - PubMed Abstract:
Venoms are excellent model systems for studying evolutionary processes associated with predator-prey interactions. Here, we present the discovery of a peptide toxin, MIITX 2 -Mg1a, which is a major component of the venom of the Australian giant red bull ant Myrmecia gulosa and has evolved to mimic, both structurally and functionally, vertebrate epidermal growth factor (EGF) peptide hormones. We show that Mg1a is a potent agonist of the mammalian EGF receptor ErbB1, and that intraplantar injection in mice causes long-lasting hypersensitivity of the injected paw. These data reveal a previously undescribed venom mode of action, highlight a role for ErbB receptors in mammalian pain signaling, and provide an example of molecular mimicry driven by defensive selection pressure.
Organizational Affiliation:
Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD 4072, Australia.