Molecular basis for the enzymatic macrocyclization of multiply backbone N-methylated peptides
Matabaro, E., Song, H., Gherlone, F., Sonderegger, L., Giltrap, A., Liver, S., Gossert, A., Kunzler, M., Naismith, J.H.(2022) bioRxiv
Experimental Data Snapshot
Starting Model: experimental
View more details
(2022) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Prolyl endopeptidase | A [auth AAA], B [auth BBB], C [auth CCC], D [auth DDD] | 745 | Omphalotus olearius | Mutation(s): 0 | 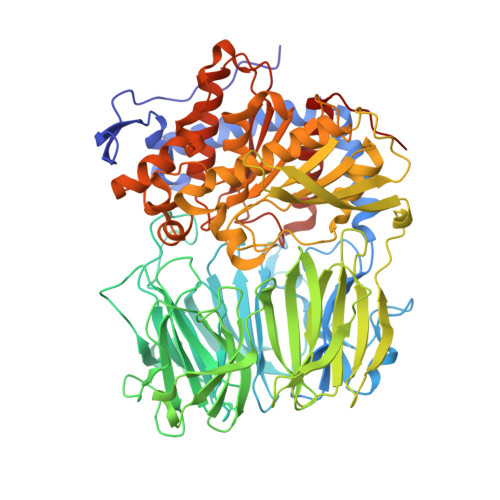 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZPR (Subject of Investigation/LOI) Query on ZPR | E [auth AAA], GA [auth CCC], KA [auth DDD], X [auth BBB] | N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL C18 H22 N2 O4 ORZXYSPOAVJYRU-HOTGVXAUSA-N |  | ||
| PO4 Query on PO4 | OA [auth DDD], RA [auth DDD] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| GOL Query on GOL | HA [auth CCC] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| DMS Query on DMS | F [auth AAA], G [auth AAA], MA [auth DDD], S [auth BBB] | DIMETHYL SULFOXIDE C2 H6 O S IAZDPXIOMUYVGZ-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | IA [auth CCC] JA [auth CCC] O [auth AAA] P [auth AAA] Q [auth AAA] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| BCT Query on BCT | AA [auth BBB] BA [auth CCC] CA [auth CCC] DA [auth CCC] EA [auth CCC] | BICARBONATE ION C H O3 BVKZGUZCCUSVTD-UHFFFAOYSA-M |  | ||
| NA Query on NA | VA [auth DDD] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_000692 (ZPR) Query on PRD_000692 | E [auth AAA], GA [auth CCC], KA [auth DDD], X [auth BBB] | Z-PRO-PROLINAL | Peptide-like / Inhibitor |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 69.68 | α = 116.23 |
| b = 102.65 | β = 101.09 |
| c = 110.27 | γ = 92.18 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| DIALS | data reduction |
| DIALS | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/R018189/1 |