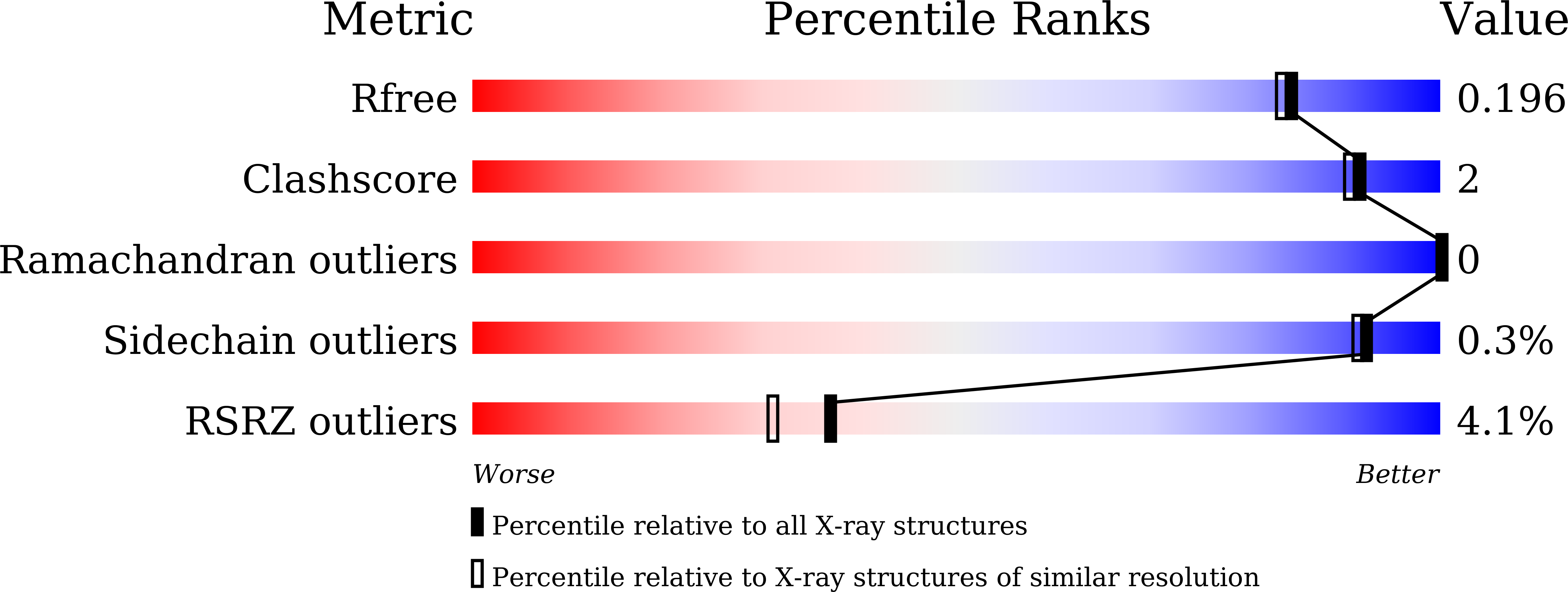
Structural insights into the PrpTA toxin-antitoxin system in Pseudoalteromonas rubra.
Wang, C., Niu, C., Hidayatullah, K.M., Xue, L., Zhu, Z., Niu, L.(2022) Front Microbiol 13: 1053255-1053255
- PubMed: 36504814
- DOI: https://doi.org/10.3389/fmicb.2022.1053255
- Primary Citation of Related Structures:
7YCS, 7YCU, 7YCV, 7YCW - PubMed Abstract:
Bacteria could survive stresses by a poorly understood mechanism that contributes to the emergence of bacterial persisters exhibiting multidrug tolerance (MDT). Recently, Pseudoalteromonas rubra prpAT module was found to encode a toxin PrpT and corresponding cognate antidote PrpA. In this study, we first reported multiple individual and complex structures of PrpA and PrpT, which uncovered the high-resolution three-dimensional structure of the PrpT:PrpA 2 :PrpT heterotetramer with the aid of size exclusion chromatography-multi-angle light scattering experiments (SEC-MALS). PrpT:PrpA 2 :PrpT is composed of a PrpA homodimer and two PrpT monomers which are relatively isolated from each other and from ParE family. The superposition of antitoxin monomer structures from these structures highlighted the flexible C-terminal domain (CTD). A striking conformational change in the CTDs of PrpA homodimer depolymerized from homotetramer was provoked upon PrpT binding, which accounts for the unique PrpT-PrpA RHH mutual interactions and further neutralizes the toxin PrpT. PrpA 2-54 -form I and II crystal structures both contain a doughnut-shaped hexadecamer formed by eight homodimers organized in a cogwheel-like form via inter-dimer interface dominated by salt bridges and hydrogen bonds. Moreover, PrpA tends to exist in solution as a homodimer other than a homotetramer (SEC-MALS) in the absence of flexible CTD. Multiple multi-dimers, tetramer and hexamer included, of PrpA 2-54 mediated by the symmetric homodimer interface and the complicated inter-dimer interface could be observed in the solution. SEC-MALS assays highlighted that phosphate buffer (PB) and the increase in the concentration appear to be favorable for the PrpA 2-54 oligomerization in the solution. Taken together with previous research, a model of PrpA 2-54 homotetramer in complex with prpAT promoter and the improved mechanism underlying how PrpTA controls the plasmid replication were proposed here.
Organizational Affiliation:
MOE Key Laboratory for Membraneless Organelles and Cellular Dynamics, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China.