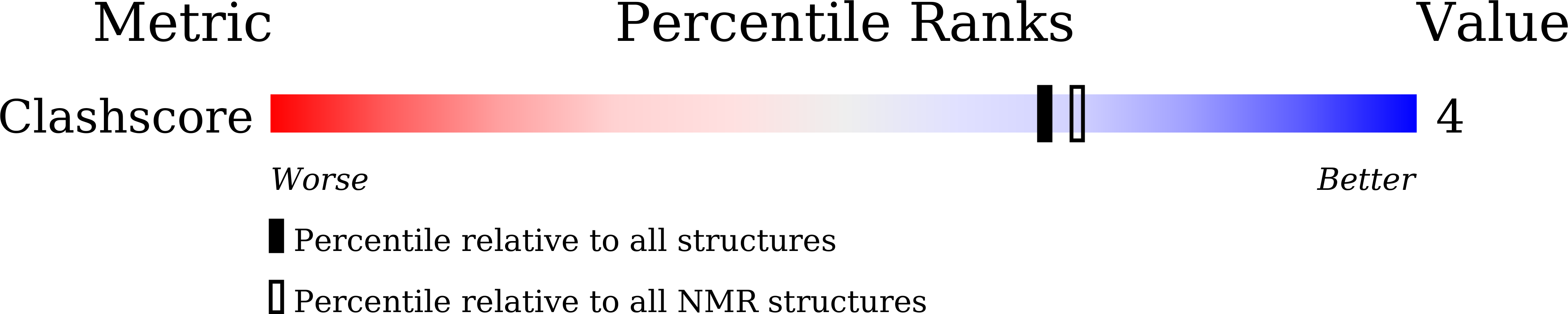Structural Basis of Pyridostatin and Its Derivatives Specifically Binding to G-Quadruplexes.
Liu, L.-Y., Ma, T.Z., Zeng, Y.L., Liu, W., Mao, Z.-W.(2022) J Am Chem Soc 144: 11878-11887
- PubMed: 35749293
- DOI: https://doi.org/10.1021/jacs.2c04775
- Primary Citation of Related Structures:
7X2Z, 7X3A - PubMed Abstract:
The nucleic acid G-quadruplex (G4) has emerged as a promising therapeutic target for a variety of diseases such as cancer and neurodegenerative disease. Among small-molecule G4-binders, pyridostatin (PDS) and its derivatives ( e.g. , PyPDS) exhibit high specificity to G4s, but the structural basis for their specific recognition of G4s remains unknown. Here, we presented two solution structures of PyPDS and PDS with a quadruplex-duplex hybrid. The structures indicate that the rigid aromatic rings of PyPDS/PDS linked by flexible amide bonds match adaptively with G-tetrad planes, enhancing π-π stacking and achieving specific recognition of G4s. The aliphatic amine side chains of PyPDS/PDS adjust conformation to interact with the phosphate backbone via hydrogen bonding and electrostatic interactions, increasing affinity for G4s. Moreover, the N-H of PyPDS/PDS amide bonds interacts with two O 6 s of G-tetrad guanines via hydrogen bonding, achieving a further increase in affinity for G4s, which is different from most G4 ligands. Our findings reveal from structural perspectives that the rational assembly of rigid and flexible structural units in a ligand can synergistically improve the selectivity and affinity for G4s through spatial selective and adaptive matching.
Organizational Affiliation:
MOE Key Laboratory of Bioinorganic and Synthetic Chemistry, School of Chemistry, Sun Yat-Sen University, Guangzhou 510275, P. R. China.