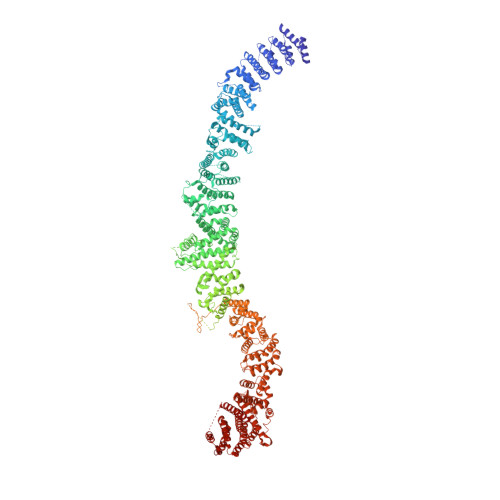
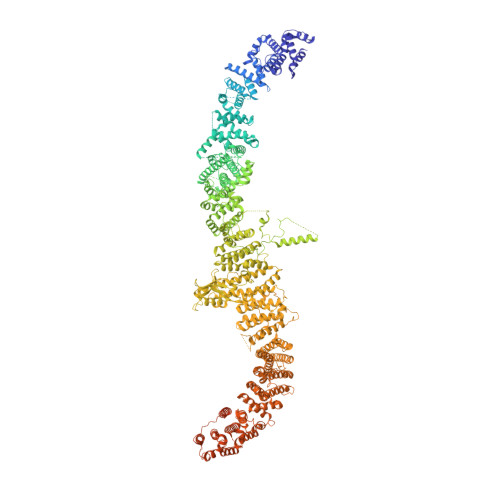
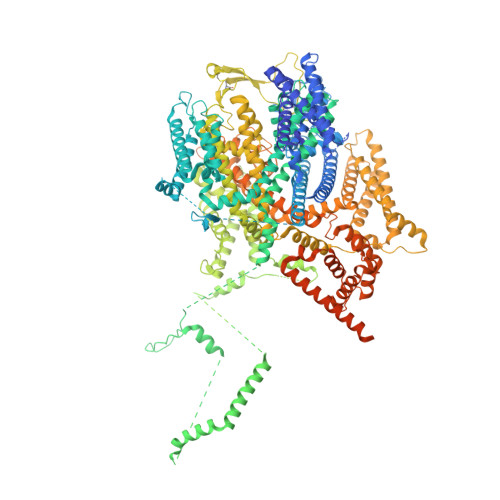
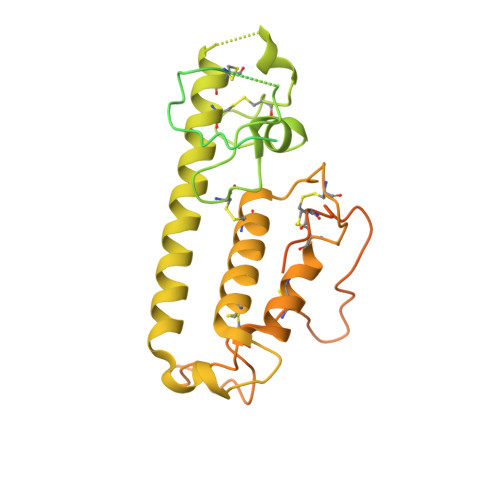
Structure and mechanism of NALCN-FAM155A-UNC79-UNC80 channel complex.
Kang, Y., Chen, L.(2022) Nat Commun 13: 2639-2639
- PubMed: 35550517
- DOI: https://doi.org/10.1038/s41467-022-30403-7
- Primary Citation of Related Structures:
7W7G - PubMed Abstract:
NALCN channel mediates sodium leak currents and is important for maintaining proper resting membrane potential. NALCN and FAM155A form the core complex of the channel, the activity of which essentially depends on the presence of both UNC79 and UNC80, two auxiliary proteins. NALCN, FAM155A, UNC79, and UNC80 co-assemble into a large hetero-tetrameric channel complex. Genetic mutations of NALCN channel components lead to neurodevelopmental diseases. However, the structure and mechanism of the intact channel complex remain elusive. Here, we present the cryo-EM structure of the mammalian NALCN-FAM155A-UNC79-UNC80 quaternary complex. The structure shows that UNC79-UNC80 form a large piler-shaped heterodimer which was tethered to the intracellular side of the NALCN channel through tripartite interactions with the cytoplasmic loops of NALCN. Two interactions are essential for proper cell surface localization of NALCN. The other interaction relieves the self-inhibition of NALCN by pulling the auto-inhibitory CTD Interacting Helix (CIH) out of its binding site. Our work defines the structural mechanism of NALCN modulation by UNC79 and UNC80.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, College of Future Technology, Institute of Molecular Medicine, Peking. University, Beijing Key Laboratory of Cardiometabolic Molecular Medicine, Beijing, 100871, China.