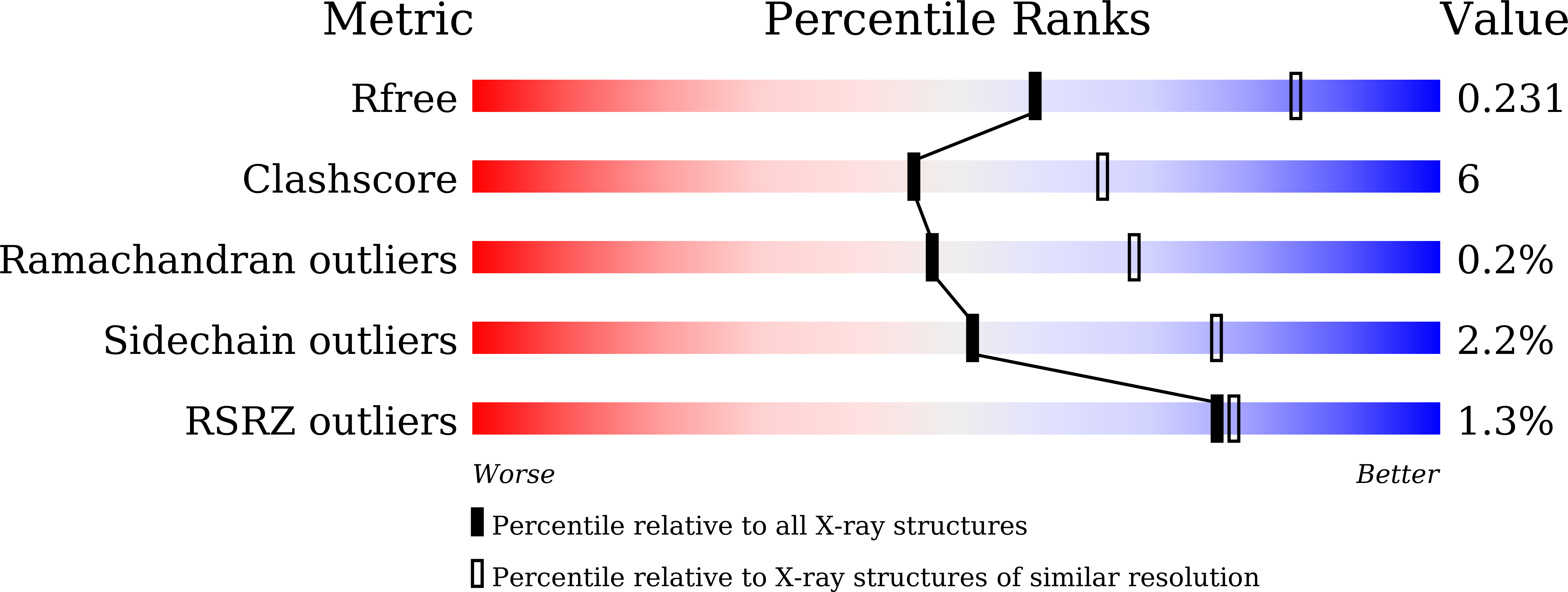
Crystal structures of glycoside hydrolase family 136 lacto-N-biosidases from monkey gut- and human adult gut bacteria.
Yamada, C., Katayama, T., Fushinobu, S.(2022) Biosci Biotechnol Biochem 86: 464-475
- PubMed: 35092420
- DOI: https://doi.org/10.1093/bbb/zbac015
- Primary Citation of Related Structures:
7V6I, 7V6M - PubMed Abstract:
Glycoside hydrolase family 136 (GH136) was established after the discovery and structural analysis of lacto-N-biosidase (LNBase) from the infant gut bacterium Bifidobacterium longum subsp. longum JCM1217 (BlLnbX). Homologous genes of BlLnbX are widely distributed in the genomes of human gut bacteria and monkey Bifidobacterium spp., although only 2 crystal structures were reported in the GH136 family. Cell suspensions of Bifidobacterium saguini, Tyzzerella nexilis, and Ruminococcus lactaris exhibited the LNBase activity. Recombinant LNBases of these 3 species were functionally expressed with their specific chaperones in Escherichia coli, and their kinetic parameters against p-nitrophenol substrates were determined. The crystal structures of the LNBases from B. saguini and T. nexilis in complex with lacto-N-biose I were determined at 2.51 and 1.92 Å resolutions, respectively. These structures conserve a β-helix fold characteristic of GH136 and the catalytic residues, but they lack the metal ions that were present in BlLnbX.
Organizational Affiliation:
Department of Biotechnology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan.