Targeting of microvillus protein Eps8 by the NleH effector kinases from enteropathogenic E. coli
Pollock, G.L., Grishin, A.M., Giogha, C., Gan, J., Oates, C.V., McMillan, P.J., Gaeta, I., Tyska, M.J., Pearson, J.S., Scott, N.E., Cygler, M., Hartland, E.L.(2022) Proc Natl Acad Sci U S A 119: e2204332119-e2204332119
- PubMed: 35976880
- DOI: https://doi.org/10.1073/pnas.2204332119
- Primary Citation of Related Structures:
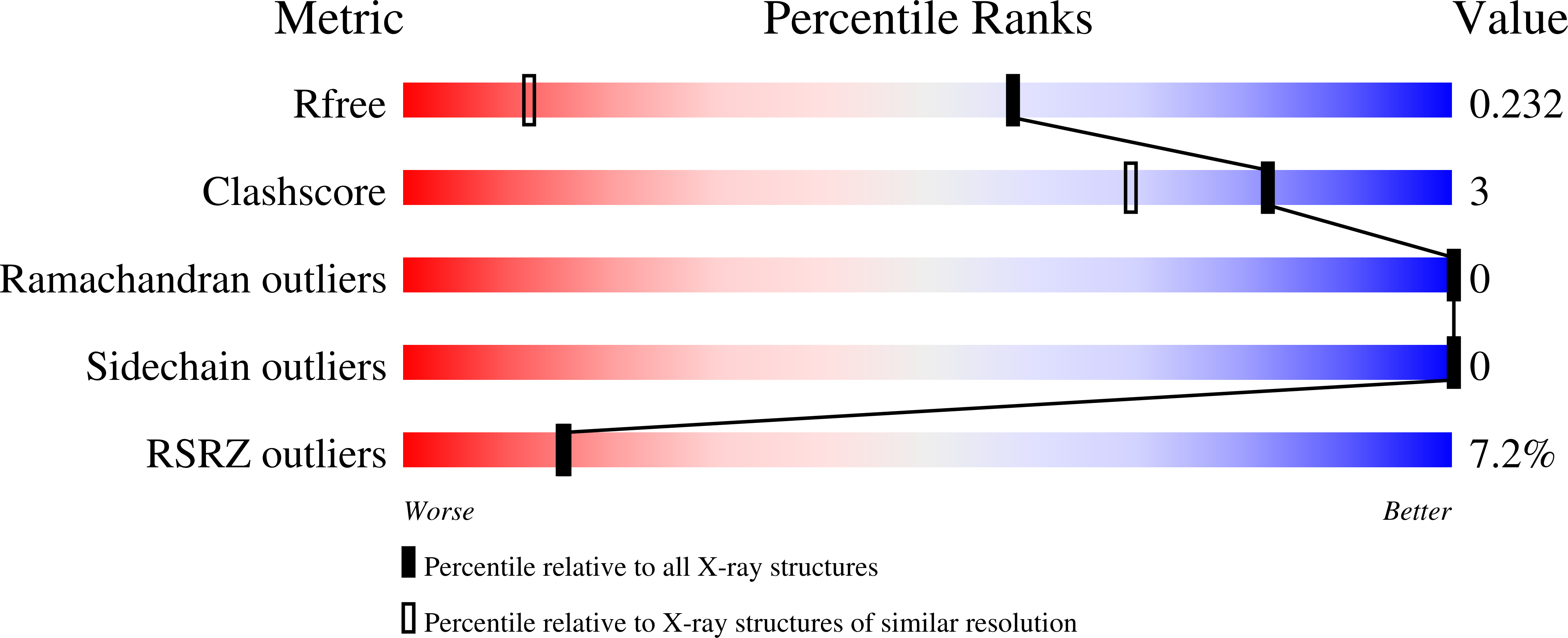
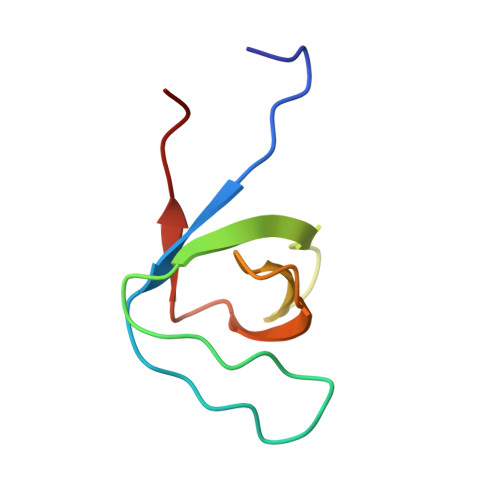
7TZK - PubMed Abstract:
Attaching and effacing (AE) lesion formation on enterocytes by enteropathogenic Escherichia coli (EPEC) requires the EPEC type III secretion system (T3SS). Two T3SS effectors injected into the host cell during infection are the atypical kinases, NleH1 and NleH2. However, the host targets of NleH1 and NleH2 kinase activity during infection have not been reported. Here phosphoproteomics identified Ser775 in the microvillus protein Eps8 as a bona fide target of NleH1 and NleH2 phosphorylation. Both kinases interacted with Eps8 through previously unrecognized, noncanonical "proline-rich" motifs, PxxDY, that bound the Src Homology 3 (SH3) domain of Eps8. Structural analysis of the Eps8 SH3 domain bound to a peptide containing one of the proline-rich motifs from NleH showed that the N-terminal part of the peptide adopts a type II polyproline helix, and its C-terminal "DY" segment makes multiple contacts with the SH3 domain. Ser775 phosphorylation by NleH1 or NleH2 hindered Eps8 bundling activity and drove dispersal of Eps8 from the AE lesion during EPEC infection. This finding suggested that NleH1 and NleH2 altered the cellular localization of Eps8 and the cytoskeletal composition of AE lesions during EPEC infection.
Organizational Affiliation:
Centre for Innate Immunity and Infectious Diseases, Hudson Institute of Medical Research, Clayton, VIC 3168, Australia.