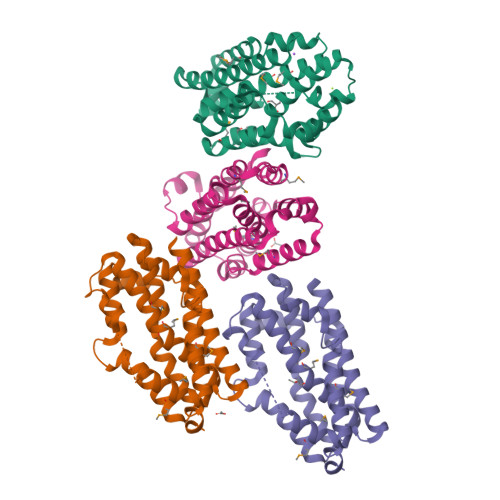
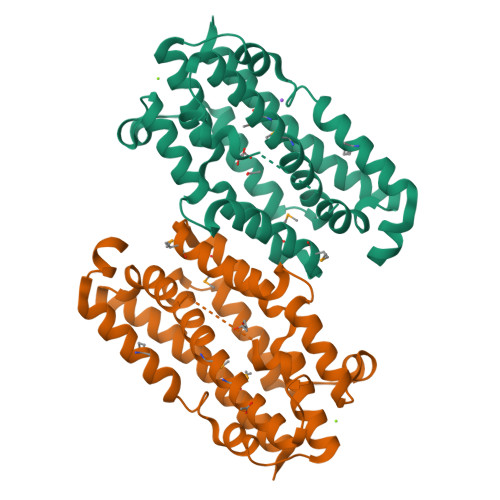
Substrate-Triggered mu-Peroxodiiron(III) Intermediate in the 4-Chloro-l-Lysine-Fragmenting Heme-Oxygenase-like Diiron Oxidase (HDO) BesC: Substrate Dissociation from, and C4 Targeting by, the Intermediate.
McBride, M.J., Nair, M.A., Sil, D., Slater, J.W., Neugebauer, M.E., Chang, M.C.Y., Boal, A.K., Krebs, C., Bollinger Jr., J.M.(2022) Biochemistry 61: 689-702
- PubMed: 35380785
- DOI: https://doi.org/10.1021/acs.biochem.1c00774
- Primary Citation of Related Structures:
7TWA - PubMed Abstract:
The enzyme BesC from the β - e thynyl-l- s erine biosynthetic pathway in Streptomyces cattleya fragments 4-chloro-l-lysine (produced from l-Lysine by BesD) to ammonia, formaldehyde, and 4-chloro-l-allylglycine and can analogously fragment l-Lys itself. BesC belongs to the emerging family of O 2 -activating non-heme-diiron enzymes with the "heme-oxygenase-like" protein fold (HDOs). Here, we show that the binding of l-Lys or an analogue triggers capture of O 2 by the protein's diiron(II) cofactor to form a blue μ-peroxodiiron(III) intermediate analogous to those previously characterized in two other HDOs, the olefin-installing fatty acid decarboxylase, UndA, and the guanidino- N -oxygenase domain of SznF. The ∼5- and ∼30-fold faster decay of the intermediate in reactions with 4-thia-l-Lys and (4 RS )-chloro-dl-lysine than in the reaction with l-Lys itself and the primary deuterium kinetic isotope effects (D-KIEs) on decay of the intermediate and production of l-allylglycine in the reaction with 4,4,5,5-[ 2 H 4 ]-l-Lys suggest that the peroxide intermediate or a reversibly connected successor complex abstracts a hydrogen atom from C4 to enable olefin formation. Surprisingly, the sluggish substrate l-Lys can dissociate after triggering intermediate formation, thereby allowing one of the better substrates to bind and react. The structure of apo BesC and the demonstrated linkage between Fe(II) and substrate binding suggest that the triggering event involves an induced ordering of ligand-providing helix 3 (α3) of the conditionally stable HDO core. As previously suggested for SznF, the dynamic α3 also likely initiates the spontaneous degradation of the diiron(III) product cluster after decay of the peroxide intermediate, a trait emerging as characteristic of the nascent HDO family.
Organizational Affiliation:
Department of Chemistry, The Pennsylvania State University, University Park, Pennsylvania 16802, United States.