Development of Selective Phosphatidylinositol 5-Phosphate 4-Kinase gamma Inhibitors with a Non-ATP-competitive, Allosteric Binding Mode.
Boffey, H.K., Rooney, T.P.C., Willems, H.M.G., Edwards, S., Green, C., Howard, T., Ogg, D., Romero, T., Scott, D.E., Winpenny, D., Duce, J., Skidmore, J., Clarke, J.H., Andrews, S.P.(2022) J Med Chem 65: 3359-3370
- PubMed: 35148092
- DOI: https://doi.org/10.1021/acs.jmedchem.1c01819
- Primary Citation of Related Structures:
7QIE, 7QPN - PubMed Abstract:
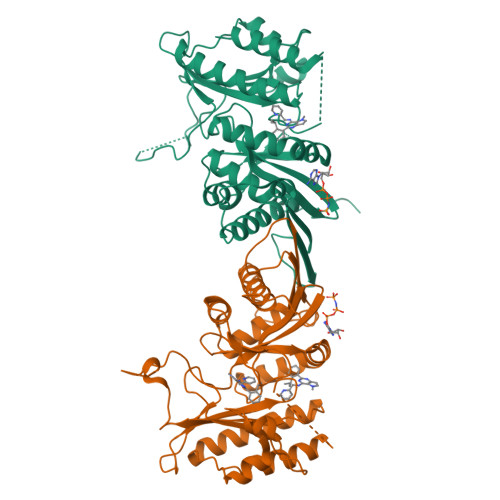
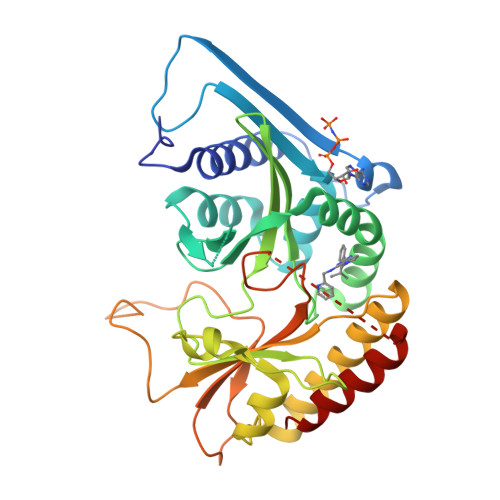
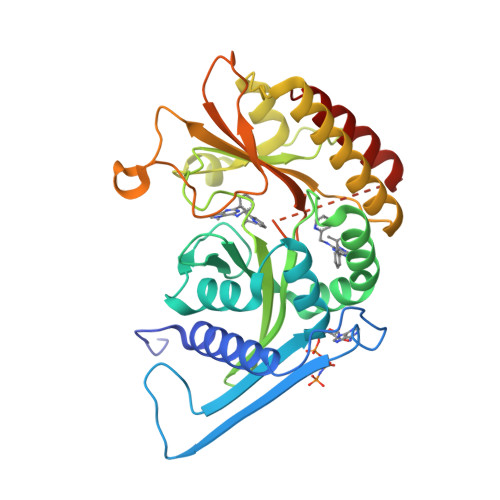
Phosphatidylinositol 5-phosphate 4-kinases (PI5P4Ks) are emerging as attractive therapeutic targets in diseases, such as cancer, immunological disorders, and neurodegeneration, owing to their central role in regulating cell signaling pathways that are either dysfunctional or can be modulated to promote cell survival. Different modes of binding may enhance inhibitor selectivity and reduce off-target effects in cells. Here, we describe efforts to improve the physicochemical properties of the selective PI5P4Kγ inhibitor, NIH-12848 ( 1 ). These improvements enabled the demonstration that this chemotype engages PI5P4Kγ in intact cells and that compounds from this series do not inhibit PI5P4Kα or PI5P4Kβ. Furthermore, the first X-ray structure of PI5P4Kγ bound to an inhibitor has been determined with this chemotype, confirming an allosteric binding mode. An exemplar from this chemical series adopted two distinct modes of inhibition, including through binding to a putative lipid interaction site which is 18 Å from the ATP pocket.
Organizational Affiliation:
The ALBORADA Drug Discovery Institute, University of Cambridge, Island Research Building, Cambridge Biomedical Campus, Hills Road, Cambridge CB2 0AH, U.K.