Structural diversity and substrate preferences of three tannase enzymes encoded by the anaerobic bacterium Clostridium butyricum.
Ristinmaa, A.S., Coleman, T., Cesar, L., Langborg Weinmann, A., Mazurkewich, S., Branden, G., Hasani, M., Larsbrink, J.(2022) J Biol Chem 298: 101758-101758
- PubMed: 35202648
- DOI: https://doi.org/10.1016/j.jbc.2022.101758
- Primary Citation of Related Structures:
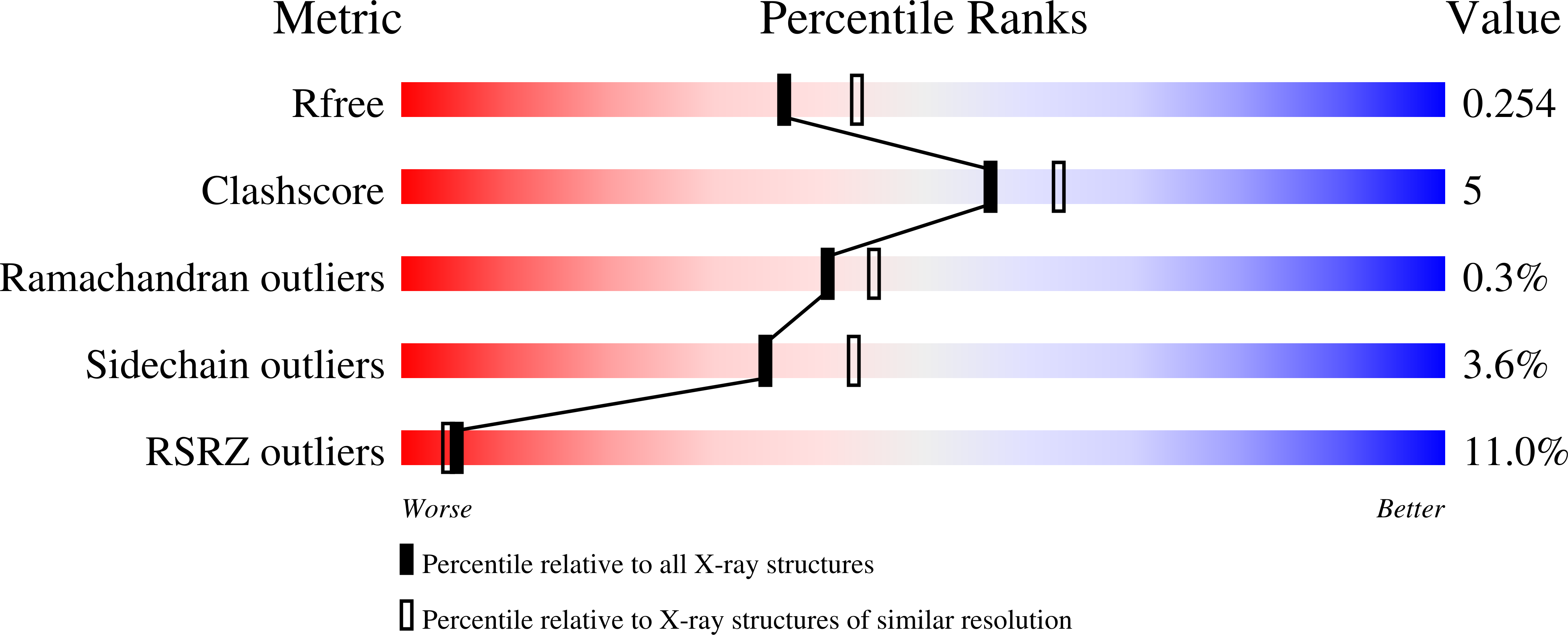
7Q6Y - PubMed Abstract:
Tannins are secondary metabolites that are enriched in the bark, roots, and knots in trees and are known to hinder microbial attack. The biological degradation of water-soluble gallotannins, such as tannic acid, is initiated by tannase enzymes (EC 3.1.1.20), which are esterases able to liberate gallic acid from aromatic-sugar complexes. However, only few tannases have previously been studied in detail. Here, for the first time, we biochemically and structurally characterize three tannases from a single organism, the anaerobic bacterium Clostridium butyricum, which inhabits both soil and gut environments. The enzymes were named CbTan1-3, and we show that each one exhibits a unique substrate preference on a range of galloyl ester model substrates; CbTan1 and 3 demonstrated preference toward galloyl esters linked to glucose, while CbTan2 was more promiscuous. All enzymes were also active on oak bark extractives. Furthermore, we solved the crystal structure of CbTan2 and produced homology models for CbTan1 and 3. In each structure, the catalytic triad and gallate-binding regions in the core domain were found in very similar positions in the active site compared with other bacterial tannases, suggesting a similar mechanism of action among these enzymes, though large inserts in each enzyme showcase overall structural diversity. In conclusion, the varied structural features and substrate specificities of the C. butyricum tannases indicate that they have different biological roles and could further be used in development of new valorization strategies for renewable plant biomass.
Organizational Affiliation:
Division of Industrial Biotechnology, Department of Biology and Biological Engineering, Chalmers University of Technology, Gothenburg, Sweden.