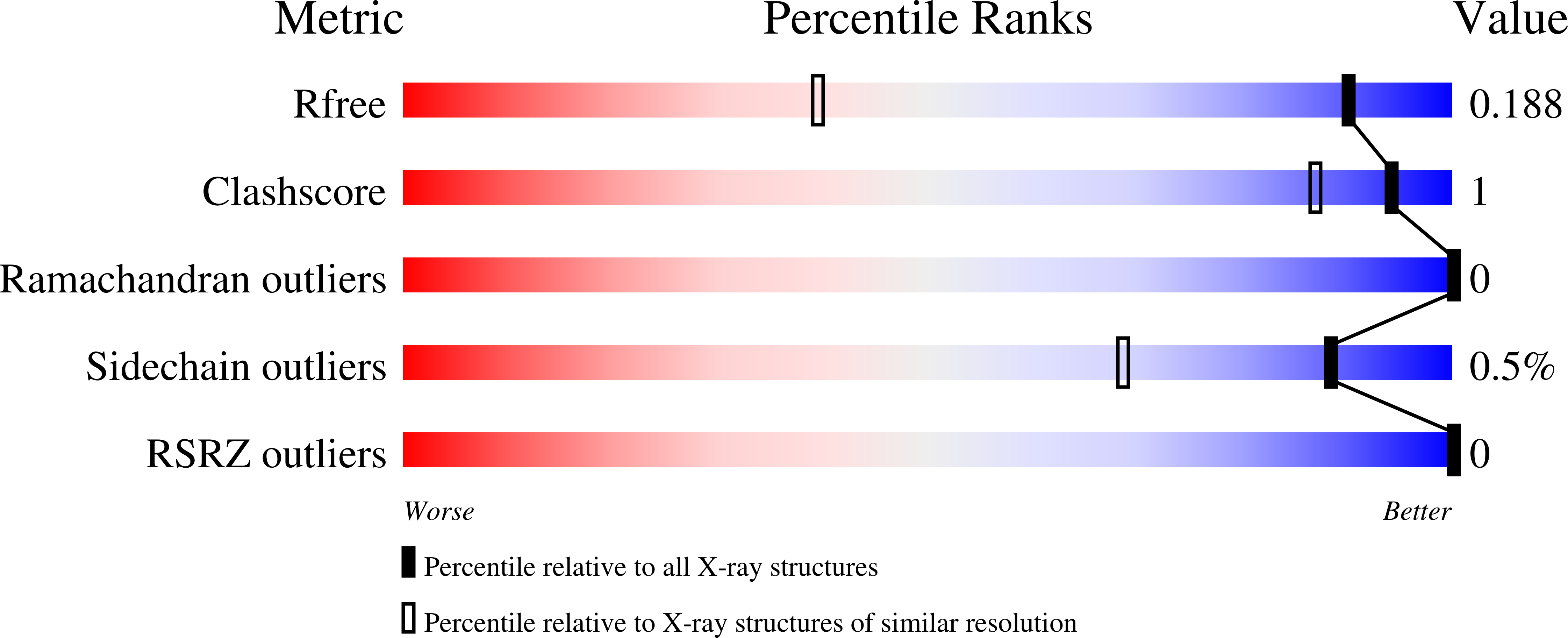
Molecular basis for the initiation of DNA primer synthesis.
Li, A.W.H., Zabrady, K., Bainbridge, L.J., Zabrady, M., Naseem-Khan, S., Berger, M.B., Kolesar, P., Cisneros, G.A., Doherty, A.J.(2022) Nature 605: 767-773
- PubMed: 35508653
- DOI: https://doi.org/10.1038/s41586-022-04695-0
- Primary Citation of Related Structures:
7NQD, 7NQE, 7NQF, 7P9J, 7QAZ - PubMed Abstract:
During the initiation of DNA replication, oligonucleotide primers are synthesized de novo by primases and are subsequently extended by replicative polymerases to complete genome duplication. The primase-polymerase (Prim-Pol) superfamily is a diverse grouping of primases, which includes replicative primases and CRISPR-associated primase-polymerases (CAPPs) involved in adaptive immunity 1-3 . Although much is known about the activities of these enzymes, the precise mechanism used by primases to initiate primer synthesis has not been elucidated. Here we identify the molecular bases for the initiation of primer synthesis by CAPP and show that this mechanism is also conserved in replicative primases. The crystal structure of a primer initiation complex reveals how the incoming nucleotides are positioned within the active site, adjacent to metal cofactors and paired to the templating single-stranded DNA strand, before synthesis of the first phosphodiester bond. Furthermore, the structure of a Prim-Pol complex with double-stranded DNA shows how the enzyme subsequently extends primers in a processive polymerase mode. The structural and mechanistic studies presented here establish how Prim-Pol proteins instigate primer synthesis, revealing the requisite molecular determinants for primer synthesis within the catalytic domain. This work also establishes that the catalytic domain of Prim-Pol enzymes, including replicative primases, is sufficient to catalyse primer formation.
Organizational Affiliation:
Genome Damage and Stability Centre, School of Life Sciences, University of Sussex, Brighton, UK.