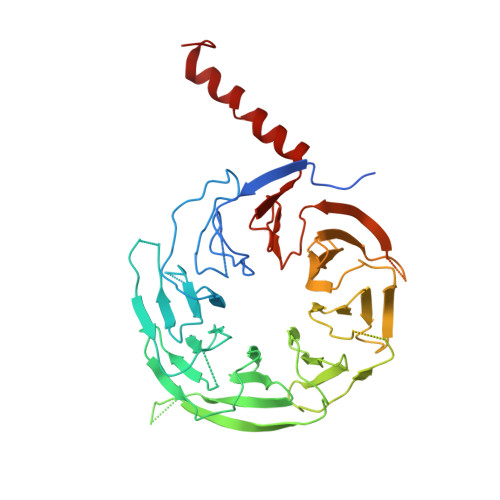
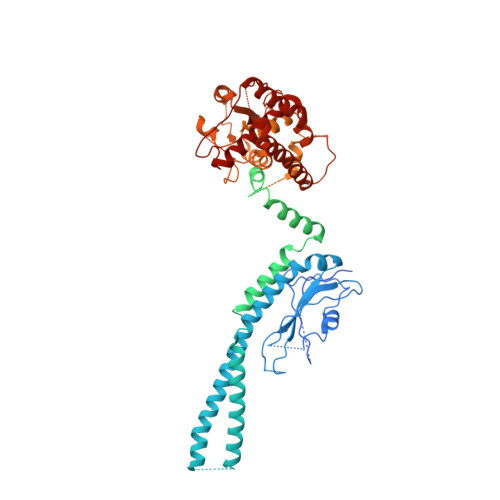
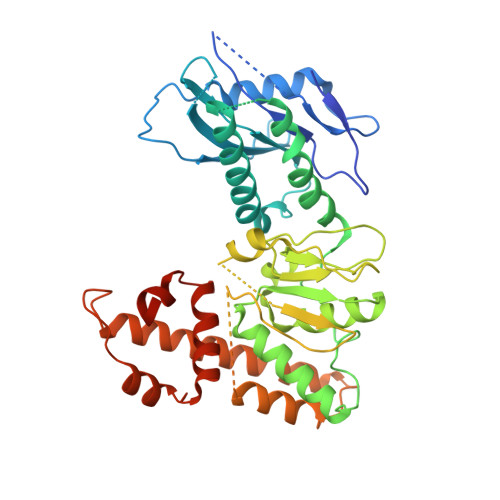
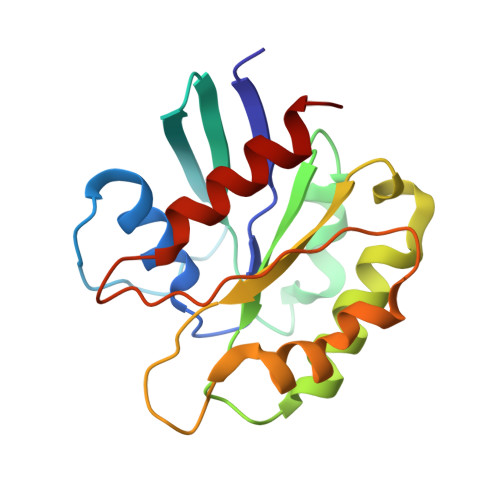
Structural basis for the ARF GAP activity and specificity of the C9orf72 complex.
Su, M.Y., Fromm, S.A., Remis, J., Toso, D.B., Hurley, J.H.(2021) Nat Commun 12: 3786-3786
- PubMed: 34145292
- DOI: https://doi.org/10.1038/s41467-021-24081-0
- Primary Citation of Related Structures:
7MGE - PubMed Abstract:
Mutation of C9ORF72 is the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontal temporal degeneration (FTD), which is attributed to both a gain and loss of function. C9orf72 forms a complex with SMCR8 and WDR41, which was reported to have GTPase activating protein activity toward ARF proteins, RAB8A, and RAB11A. We determined the cryo-EM structure of ARF1-GDP-BeF 3 - bound to C9orf72:SMCR8:WDR41. The SMCR8 longin and C9orf72 longin domains form the binding pocket for ARF1. One face of the C9orf72 longin domain holds ARF1 in place, while the SMCR8 longin positions the catalytic finger Arg147 in the ARF1 active site. Mutations in interfacial residues of ARF1 and C9orf72 reduced or eliminated GAP activity. RAB8A GAP required ~10-fold higher concentrations of the C9orf72 complex than for ARF1. These data support a specific function for the C9orf72 complex as an ARF GAP. The structure also provides a model for the active forms of the longin domain GAPs of FLCN and NPRL2 that regulate the Rag GTPases of the mTORC1 pathway.
Organizational Affiliation:
School of Medicine, Southern University of Science and Technology, Shenzhen, China.