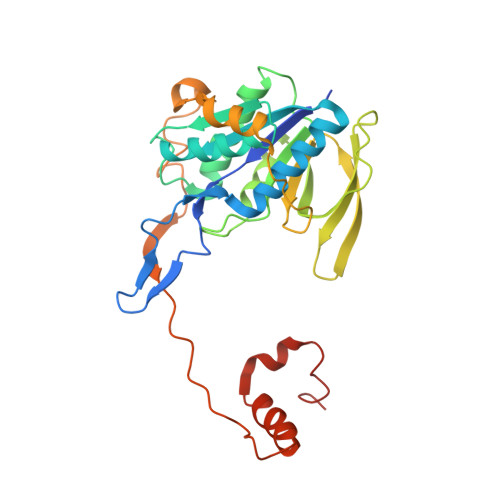
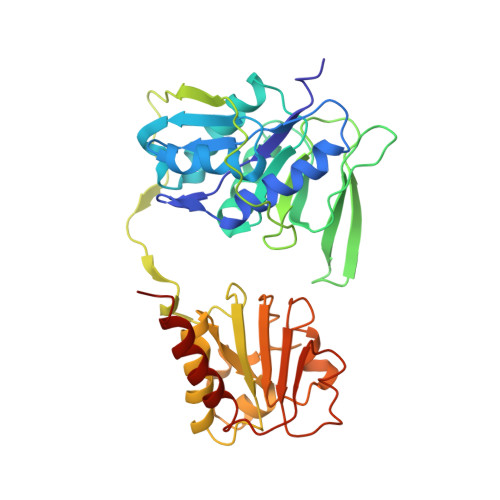
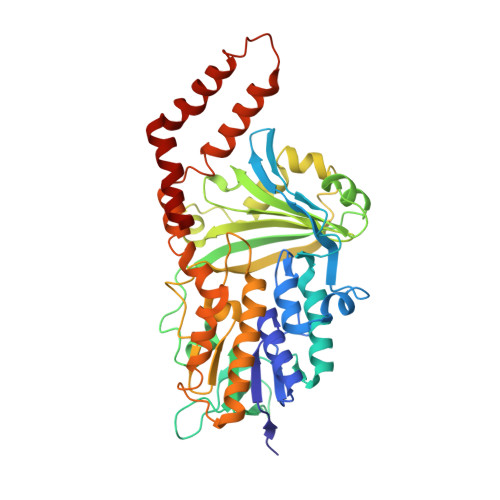
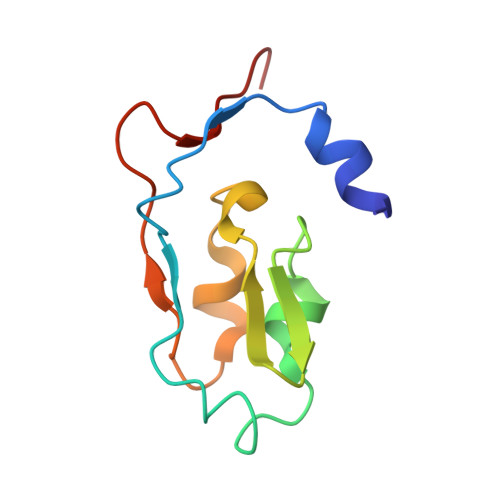
Cryoelectron microscopy structure and mechanism of the membrane-associated electron-bifurcating flavoprotein Fix/EtfABCX.
Feng, X., Schut, G.J., Lipscomb, G.L., Li, H., Adams, M.W.W.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33372143
- DOI: https://doi.org/10.1073/pnas.2016978118
- Primary Citation of Related Structures:
7KOE - PubMed Abstract:
The electron-transferring flavoprotein-menaquinone oxidoreductase ABCX (EtfABCX), also known as FixABCX for its role in nitrogen-fixing organisms, is a member of a family of electron-transferring flavoproteins that catalyze electron bifurcation. EtfABCX enables endergonic reduction of ferredoxin ( E °' ∼-450 mV) using NADH ( E °' -320 mV) as the electron donor by coupling this reaction to the exergonic reduction of menaquinone ( E °' -80 mV). Here we report the 2.9 Å structure of EtfABCX, a membrane-associated flavin-based electron bifurcation (FBEB) complex, from a thermophilic bacterium. EtfABCX forms a superdimer with two membrane-associated EtfCs at the dimer interface that contain two bound menaquinones. The structure reveals that, in contrast to previous predictions, the low-potential electrons bifurcated from EtfAB are most likely directly transferred to ferredoxin, while high-potential electrons reduce the quinone via two [4Fe-4S] clusters in EtfX. Surprisingly, EtfX shares remarkable structural similarity with mammalian [4Fe-4S] cluster-containing ETF ubiquinone oxidoreductase (ETF-QO), suggesting an unexpected evolutionary link between bifurcating and nonbifurcating systems. Based on this structure and spectroscopic studies of a closely related EtfABCX, we propose a detailed mechanism of the catalytic cycle and the accompanying structural changes in this membrane-associated FBEB system.
Organizational Affiliation:
Department of Structural Biology, Van Andel Institute, Grand Rapids, MI 49503.