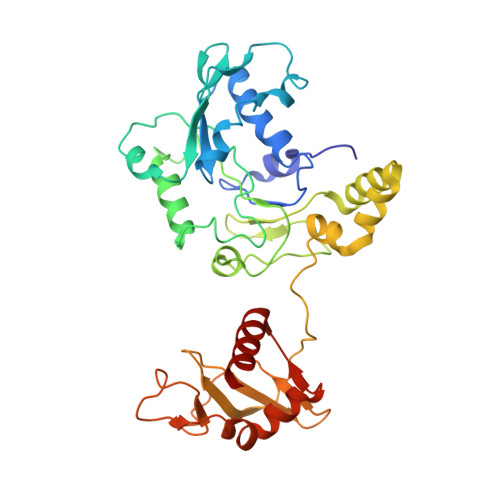
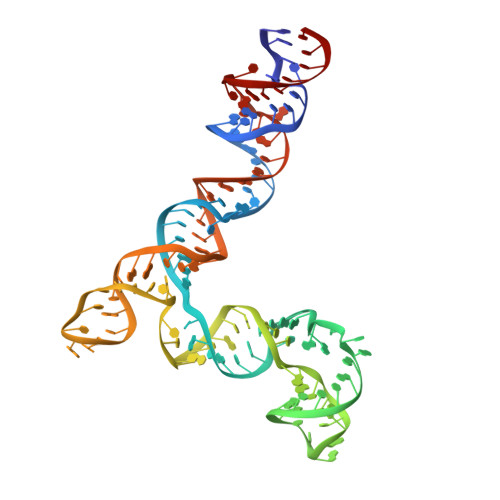
A viral genome packaging motor transitions between cyclic and helical symmetry to translocate dsDNA.
Woodson, M., Pajak, J., Mahler, B.P., Zhao, W., Zhang, W., Arya, G., White, M.A., Jardine, P.J., Morais, M.C.(2021) Sci Adv 7
- PubMed: 33962953
- DOI: https://doi.org/10.1126/sciadv.abc1955
- Primary Citation of Related Structures:
7JQQ - PubMed Abstract:
Molecular segregation and biopolymer manipulation require the action of molecular motors to do work by applying directional forces to macromolecules. The additional strand conserved E (ASCE) ring motors are an ancient family of molecular motors responsible for diverse biological polymer manipulation tasks. Viruses use ASCE segregation motors to package their genomes into their protein capsids and provide accessible experimental systems due to their relative simplicity. We show by cryo-EM-focused image reconstruction that ASCE ATPases in viral double-stranded DNA (dsDNA) packaging motors adopt helical symmetry complementary to their dsDNA substrates. Together with previous data, our results suggest that these motors cycle between helical and planar configurations, providing a possible mechanism for directional translocation of DNA. Similar changes in quaternary structure have been observed for proteasome and helicase motors, suggesting an ancient and common mechanism of force generation that has been adapted for specific tasks over the course of evolution.
Organizational Affiliation:
Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, Galveston, TX 77555, USA.