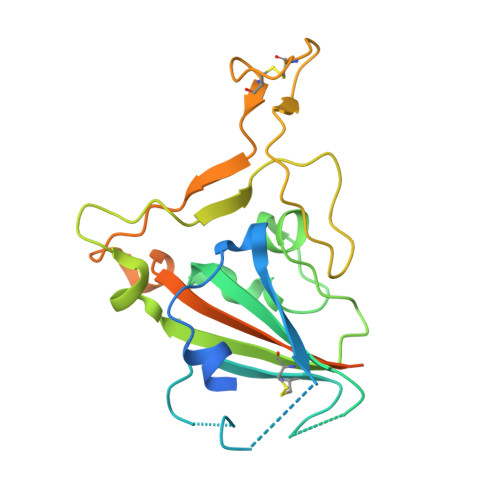
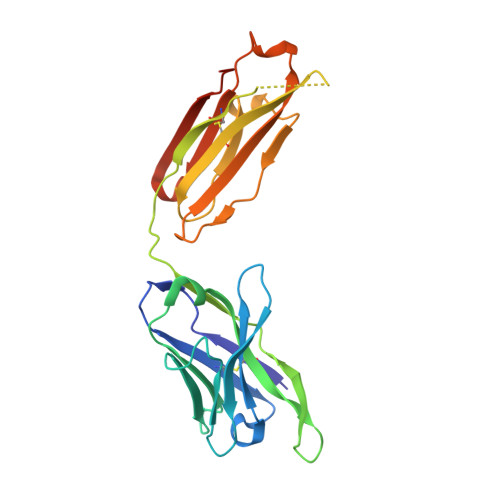
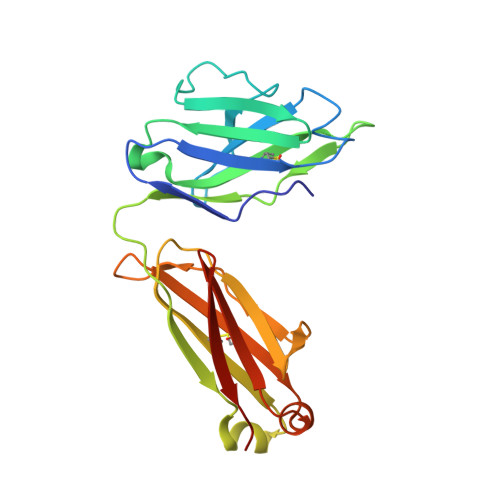
An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain.
Wu, N.C., Yuan, M., Liu, H., Lee, C.D., Zhu, X., Bangaru, S., Torres, J.L., Caniels, T.G., Brouwer, P.J.M., van Gils, M.J., Sanders, R.W., Ward, A.B., Wilson, I.A.(2020) Cell Rep 33: 108274-108274
- PubMed: 33027617
- DOI: https://doi.org/10.1016/j.celrep.2020.108274
- Primary Citation of Related Structures:
7JMO, 7JMP - PubMed Abstract:
IGHV3-53-encoded neutralizing antibodies are commonly elicited during SARS-CoV-2 infection and target the receptor-binding domain (RBD) of the spike (S) protein. Such IGHV3-53 antibodies generally have a short CDR H3 because of structural constraints in binding the RBD (mode A). However, a small subset of IGHV3-53 antibodies to the RBD contain a longer CDR H3. Crystal structures of two IGHV3-53 neutralizing antibodies here demonstrate that a longer CDR H3 can be accommodated in a different binding mode (mode B). These two classes of IGHV3-53 antibodies both target the ACE2 receptor binding site, but with very different angles of approach and molecular interactions. Overall, these findings emphasize the versatility of IGHV3-53 in this common antibody response to SARS-CoV-2, where conserved IGHV3-53 germline-encoded features can be combined with very different CDR H3 lengths and light chains for SARS-CoV-2 RBD recognition and virus neutralization.
Organizational Affiliation:
Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA; Carl R. Woese Institute for Genomic Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.