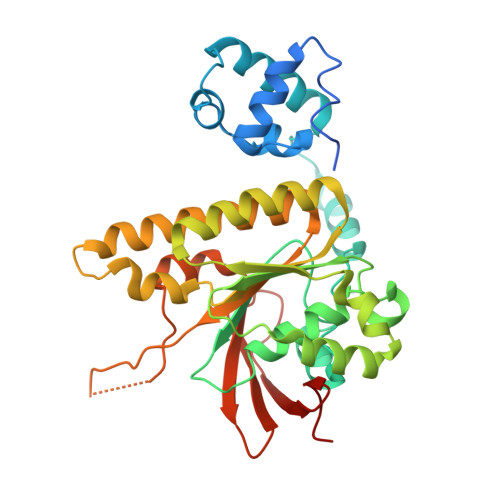
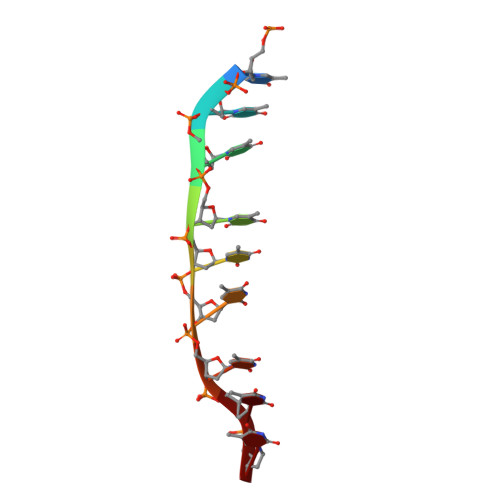
Mechanisms of distinctive mismatch tolerance between Rad51 and Dmc1 in homologous recombination.
Xu, J., Zhao, L., Peng, S., Chu, H., Liang, R., Tian, M., Connell, P.P., Li, G., Chen, C., Wang, H.W.(2021) Nucleic Acids Res 49: 13135-13149
- PubMed: 34871438
- DOI: https://doi.org/10.1093/nar/gkab1141
- Primary Citation of Related Structures:
7EJ6, 7EJ7, 7EJC, 7EJE - PubMed Abstract:
Homologous recombination (HR) is a primary DNA double-strand breaks (DSBs) repair mechanism. The recombinases Rad51 and Dmc1 are highly conserved in the RecA family; Rad51 is mainly responsible for DNA repair in somatic cells during mitosis while Dmc1 only works during meiosis in germ cells. This spatiotemporal difference is probably due to their distinctive mismatch tolerance during HR: Rad51 does not permit HR in the presence of mismatches, whereas Dmc1 can tolerate certain mismatches. Here, the cryo-EM structures of Rad51-DNA and Dmc1-DNA complexes revealed that the major conformational differences between these two proteins are located in their Loop2 regions, which contain invading single-stranded DNA (ssDNA) binding residues and double-stranded DNA (dsDNA) complementary strand binding residues, stabilizing ssDNA and dsDNA in presynaptic and postsynaptic complexes, respectively. By combining molecular dynamic simulation and single-molecule FRET assays, we identified that V273 and D274 in the Loop2 region of human RAD51 (hRAD51), corresponding to P274 and G275 of human DMC1 (hDMC1), are the key residues regulating mismatch tolerance during strand exchange in HR. This HR accuracy control mechanism provides mechanistic insights into the specific roles of Rad51 and Dmc1 in DNA double-strand break repair and may shed light on the regulatory mechanism of genetic recombination in mitosis and meiosis.
Organizational Affiliation:
Ministry of Education Key Laboratory of Protein Sciences, Tsinghua-Peking Joint Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing, 100084, China.