Crystal structure of mutant of a Petase mutant
Han, X., Liu, W.D., Zheng, Y.Y., Chen, C.C., Guo, R.T.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Poly(ethylene terephthalate) hydrolase | 277 | Piscinibacter sakaiensis | Mutation(s): 1 Gene Names: ISF6_4831 EC: 3.1.1.101 | 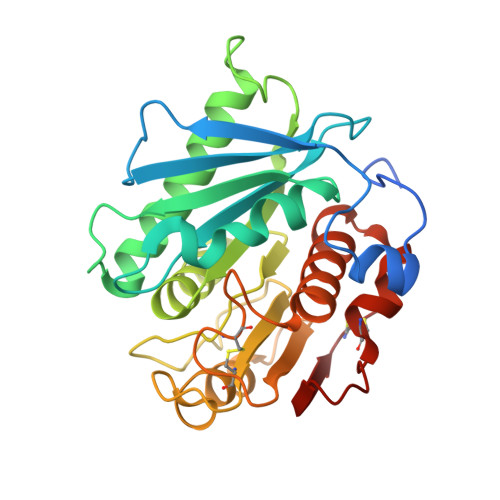 | |
UniProt | |||||
Find proteins for A0A0K8P6T7 (Ideonella sakaiensis (strain NBRC 110686 / TISTR 2288 / 201-F6)) Explore A0A0K8P6T7 Go to UniProtKB: A0A0K8P6T7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A0K8P6T7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 51.065 | α = 90 |
| b = 51.264 | β = 90 |
| c = 84.806 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data scaling |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 3180062 |
| Chinese Academy of Sciences | China | KFJ-STS-ZDTP-064 |