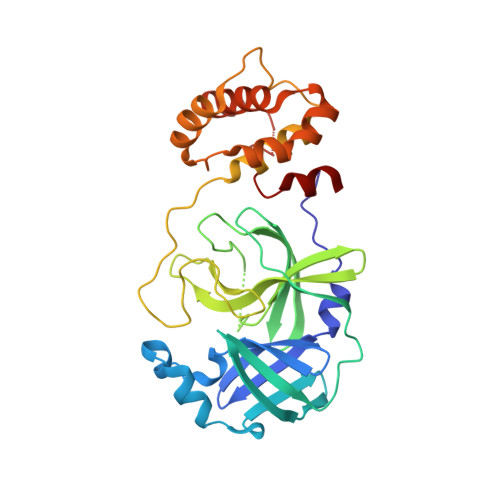
Structure of SARS-CoV-2 main protease in the apo state.
Zhou, X., Zhong, F., Lin, C., Hu, X., Zhang, Y., Xiong, B., Yin, X., Fu, J., He, W., Duan, J., Fu, Y., Zhou, H., McCormick, P.J., Wang, Q., Li, J., Zhang, J.(2021) Sci China Life Sci 64: 656-659
- PubMed: 32880863
- DOI: https://doi.org/10.1007/s11427-020-1791-3
- Primary Citation of Related Structures:
7C2Q
Organizational Affiliation:
School of Basic Medical Sciences, Nanchang University, Nanchang, 330031, China.