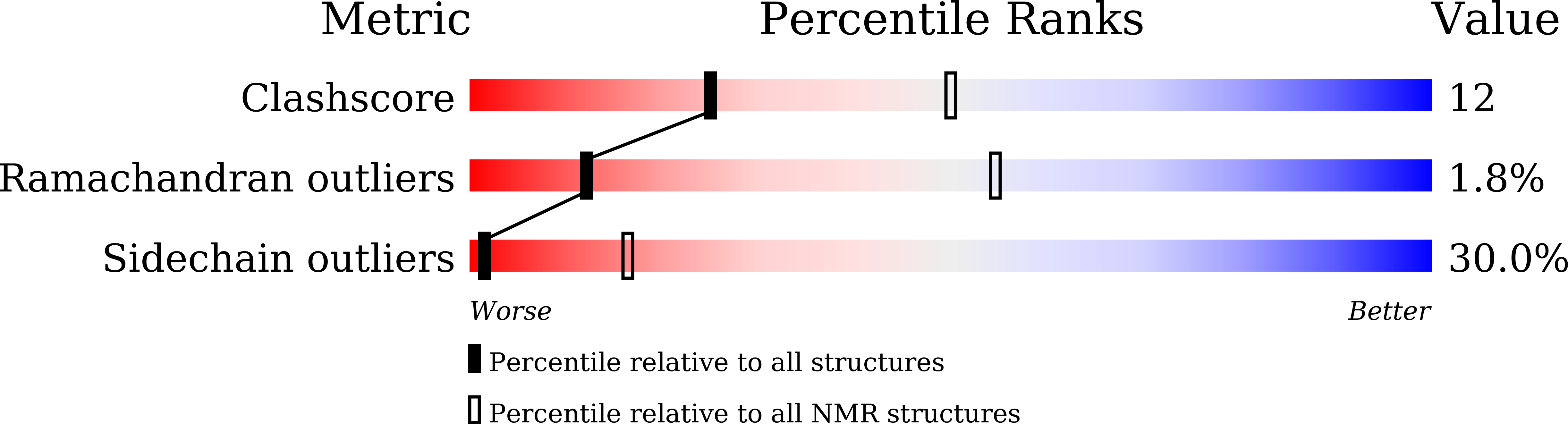Structure of a consensus chitin-binding domain revealed by solution NMR.
Heymann, D., Mohanram, H., Kumar, A., Verma, C.S., Lescar, J., Miserez, A.(2021) J Struct Biol 213: 107725-107725
- PubMed: 33744410
- DOI: https://doi.org/10.1016/j.jsb.2021.107725
- Primary Citation of Related Structures:
7BWE, 7BWO - PubMed Abstract:
Chitin-binding proteins (CBPs) are a versatile group of proteins found in almost every organism on earth. CBPs are involved in enzymatic carbohydrate degradation and also serve as templating scaffolds in the exoskeleton of crustaceans and insects. One specific chitin-binding motif found across a wide range of arthropods' exoskeletons is the "extended Rebers and Riddiford" consensus (R&R), whose mechanism of chitin binding remains unclear. Here, we report the 3D structure and molecular level interactions of a chitin-binding domain (CBD-γ) located in a CBP from the beak of the jumbo squid Dosidicus gigas. This CBP is one of four chitin-binding proteins identified in the beak mouthpart of D. gigas and is believed to interact with chitin to form a scaffold network that is infiltrated with a second set of structural proteins during beak maturation. We used solution state NMR spectroscopy to elucidate the molecular interactions between CBD-γ and the soluble chitin derivative pentaacetyl-chitopentaose (PCP), and find that folding of CBD-γ is triggered upon its interaction with PCP. To our knowledge, this is the first experimental 3D structure of a CBP containing the R&R consensus motif, which can be used as a template to understand in more details the role of the R&R motif found in a wide range of CBP-chitin complexes. The present structure also provides molecular information for biomimetic synthesis of graded biomaterials using aqueous-based chemistry and biopolymers.
Organizational Affiliation:
Biological and Biomimetic Material Laboratory, Center for Sustainable Materials (SusMat), School of Materials Science and Engineering, Nanyang Technological University (NTU), 50 Nanyang Avenue, Singapore 637553, Singapore; NTU Institute of Structural Biology, Experimental Medicine Building (EMB), 59 Nanyang Drive, Level 06-01, Singapore 636921, Singapore.