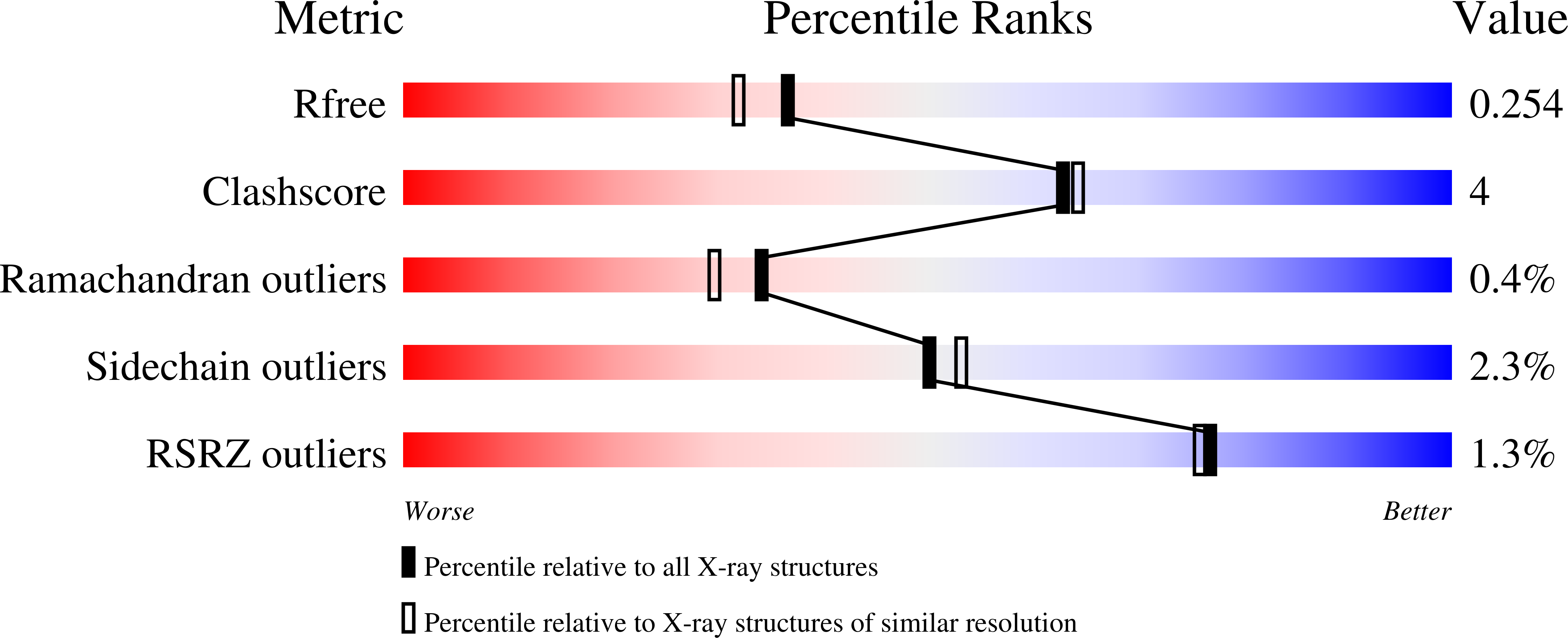
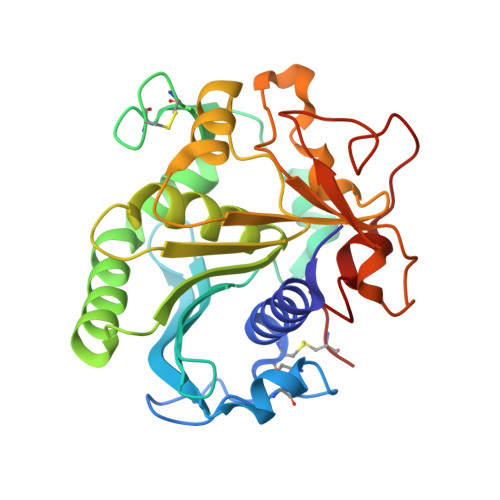
Production of Cross-Linked Lipase Crystals at a Preparative Scale.
Fernandez-Penas, R., Verdugo-Escamilla, C., Martinez-Rodriguez, S., Gavira, J.A.(2021) Cryst Growth Des 21: 1698-1707
- PubMed: 34602865
- DOI: https://doi.org/10.1021/acs.cgd.0c01608
- Primary Citation of Related Structures:
7APN, 7APP - PubMed Abstract:
The autoimmobilization of enzymes via cross-linked enzyme crystals (CLECs) has regained interest in recent years, boosted by the extensive knowledge gained in protein crystallization, the decrease of cost and laboriousness of the process, and the development of potential applications. In this work, we present the crystallization and preparative-scale production of reinforced cross-linked lipase crystals (RCLLCs) using a commercial detergent additive as a raw material. Bulk crystallization was carried out in 500 mL of agarose media using the batch technique. Agarose facilitates the homogeneous production of crystals, their cross-linking treatment, and their extraction. RCLLCs were active in an aqueous solution and in hexane, as shown by the hydrolysis of p -nitrophenol butyrate and α-methylbenzyl acetate, respectively. RCLLCs presented both high thermal and robust operational stability, allowing the preparation of a packed-bed chromatographic column to work in a continuous flow. Finally, we determined the three-dimensional (3D) models of this commercial lipase crystallized with and without phosphate at 2.0 and 1.7 Å resolutions, respectively.
Organizational Affiliation:
Laboratorio de Estudios Cristalográficos, Instituto Andaluz de Ciencias de la Tierra, Consejo Superior de Investigaciones Científicas-Universidad de Granada, Avenida de las Palmeras 4, Armilla, 18100 Granada, Spain.