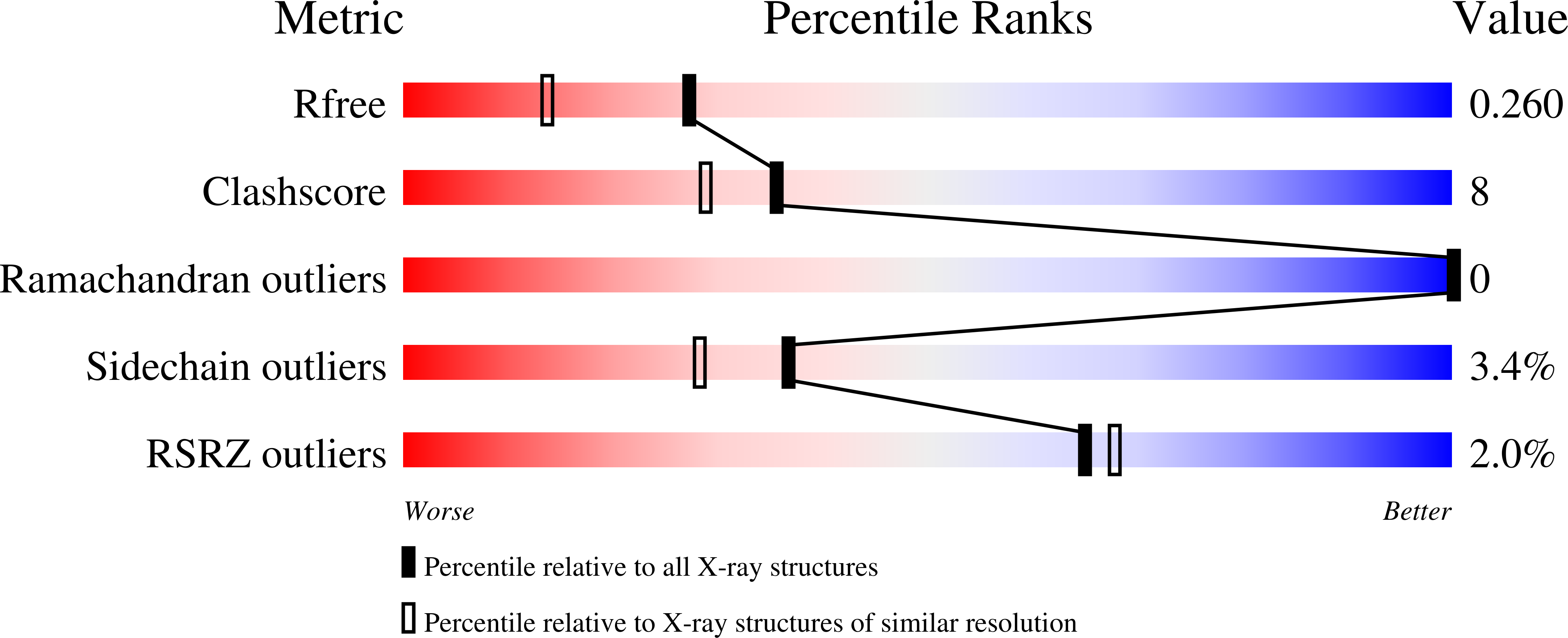
Structural characterization of KKT4, an unconventional microtubule-binding kinetochore protein.
Ludzia, P., Lowe, E.D., Marciano, G., Mohammed, S., Redfield, C., Akiyoshi, B.(2021) Structure 29: 1014-1028.e8
- PubMed: 33915106
- DOI: https://doi.org/10.1016/j.str.2021.04.004
- Primary Citation of Related Structures:
6ZPJ, 6ZPK, 6ZPM - PubMed Abstract:
The kinetochore is the macromolecular machinery that drives chromosome segregation by interacting with spindle microtubules. Kinetoplastids (such as Trypanosoma brucei), a group of evolutionarily divergent eukaryotes, have a unique set of kinetochore proteins that lack any significant homology to canonical kinetochore components. To date, KKT4 is the only kinetoplastid kinetochore protein that is known to bind microtubules. Here we use X-ray crystallography, NMR spectroscopy, and crosslinking mass spectrometry to characterize the structure and dynamics of KKT4. We show that its microtubule-binding domain consists of a coiled-coil structure followed by a positively charged disordered tail. The structure of the C-terminal BRCT domain of KKT4 reveals that it is likely a phosphorylation-dependent protein-protein interaction domain. The BRCT domain interacts with the N-terminal region of the KKT4 microtubule-binding domain and with a phosphopeptide derived from KKT8. Taken together, these results provide structural insights into the unconventional kinetoplastid kinetochore protein KKT4.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, Oxford OX1 3QU, UK.