Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Bochel, A.J., Williams, C., McCoy, A.J., Hoppe, H.J., Winter, A.J., Nicholls, R.D., Harlos, K., Jones, E.Y., Berger, I., Hassan, A.B., Crump, M.P.(2020) Structure 28: 1300-1312.e5
- PubMed: 32877646
- DOI: https://doi.org/10.1016/j.str.2020.08.002
- Primary Citation of Related Structures:
6Z30, 6Z31, 6Z32 - PubMed Abstract:
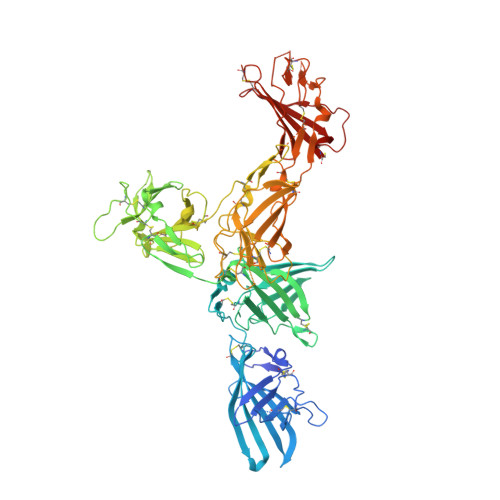
The cation-independent mannose 6-phosphate (M6P)/Insulin-like growth factor-2 receptor (CI-MPR/IGF2R) is an ∼300 kDa transmembrane protein responsible for trafficking M6P-tagged lysosomal hydrolases and internalizing IGF2. The extracellular region of the CI-MPR has 15 homologous domains, including M6P-binding domains (D) 3, 5, 9, and 15 and IGF2-binding domain 11. We have focused on solving the first structures of human D7-10 within two multi-domain constructs, D9-10 and D7-11, and provide the first high-resolution description of the high-affinity M6P-binding D9. Moreover, D9 stabilizes a well-defined hub formed by D7-11 whereby two penta-domains intertwine to form a dimeric helical-type coil via an N-glycan bridge on D9. Remarkably the D7-11 structure matches an IGF2-bound state of the receptor, suggesting this may be an intrinsically stable conformation at neutral pH. Interdomain clusters of histidine and proline residues may impart receptor rigidity and play a role in structural transitions at low pH.
Organizational Affiliation:
School of Chemistry, Cantock's Close, University of Bristol, Bristol BS8 1TS, UK.