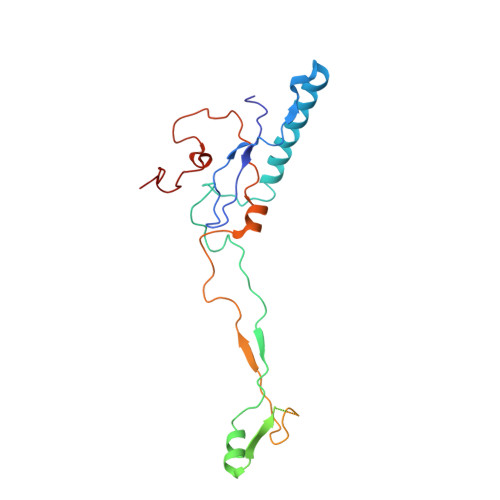
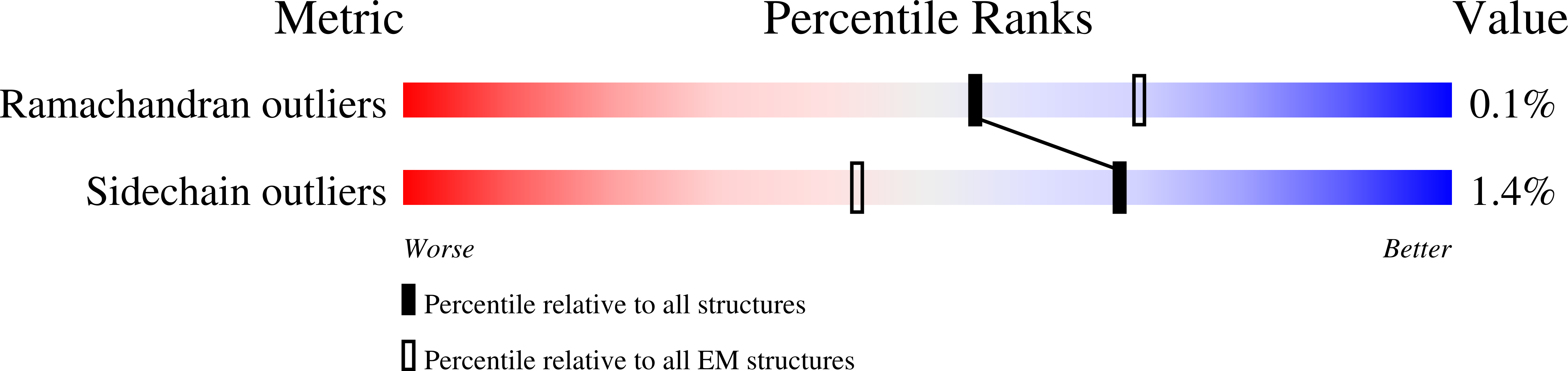Cryo-EM structure of enteric adenovirus HAdV-F41 highlights structural variations among human adenoviruses.
Perez-Illana, M., Martinez, M., Condezo, G.N., Hernando-Perez, M., Mangroo, C., Brown, M., Marabini, R., San Martin, C.(2021) Sci Adv 7
- PubMed: 33627423
- DOI: https://doi.org/10.1126/sciadv.abd9421
- Primary Citation of Related Structures:
6YBA - PubMed Abstract:
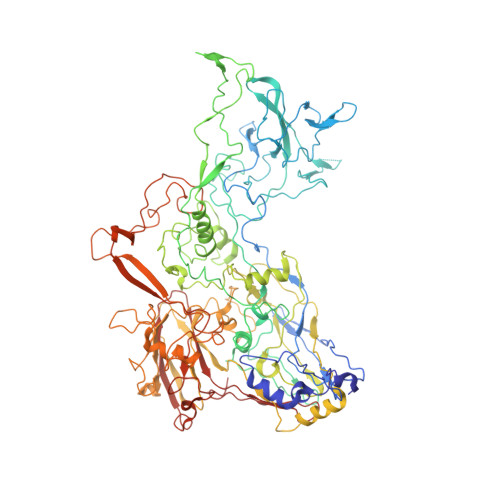
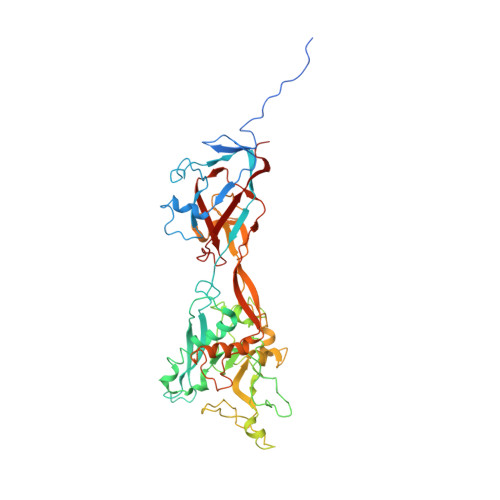
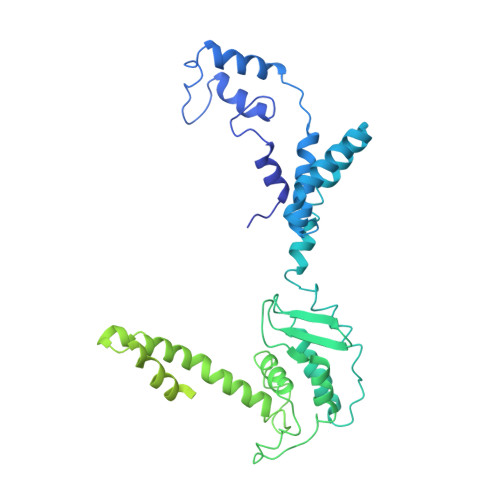
Enteric adenoviruses, one of the main causes of viral gastroenteritis in the world, must withstand the harsh conditions found in the gut. This requirement suggests that capsid stability must be different from that of other adenoviruses. We report the 4-Å-resolution structure of a human enteric adenovirus, HAdV-F41, and compare it with that of other adenoviruses with respiratory (HAdV-C5) and ocular (HAdV-D26) tropisms. While the overall structures of hexon, penton base, and internal minor coat proteins IIIa and VIII are conserved, we observe partially ordered elements reinforcing the vertex region, which suggests their role in enhancing the physicochemical capsid stability of HAdV-F41. Unexpectedly, we find an organization of the external minor coat protein IX different from all previously characterized human and nonhuman mastadenoviruses. Knowledge of the structure of enteric adenoviruses provides a starting point for the design of vectors suitable for oral delivery or intestinal targeting.
Organizational Affiliation:
Department of Macromolecular Structures, Centro Nacional de Biotecnología (CNB-CSIC), Madrid, Spain.