Functional reprogramming ofCandida glabrataepithelial adhesins: the role of conserved and variable structural motifs in ligand binding.
Hoffmann, D., Diderrich, R., Reithofer, V., Friederichs, S., Kock, M., Essen, L.O., Mosch, H.U.(2020) J Biol Chem 295: 12512-12524
- PubMed: 32669365
- DOI: https://doi.org/10.1074/jbc.RA120.013968
- Primary Citation of Related Structures:
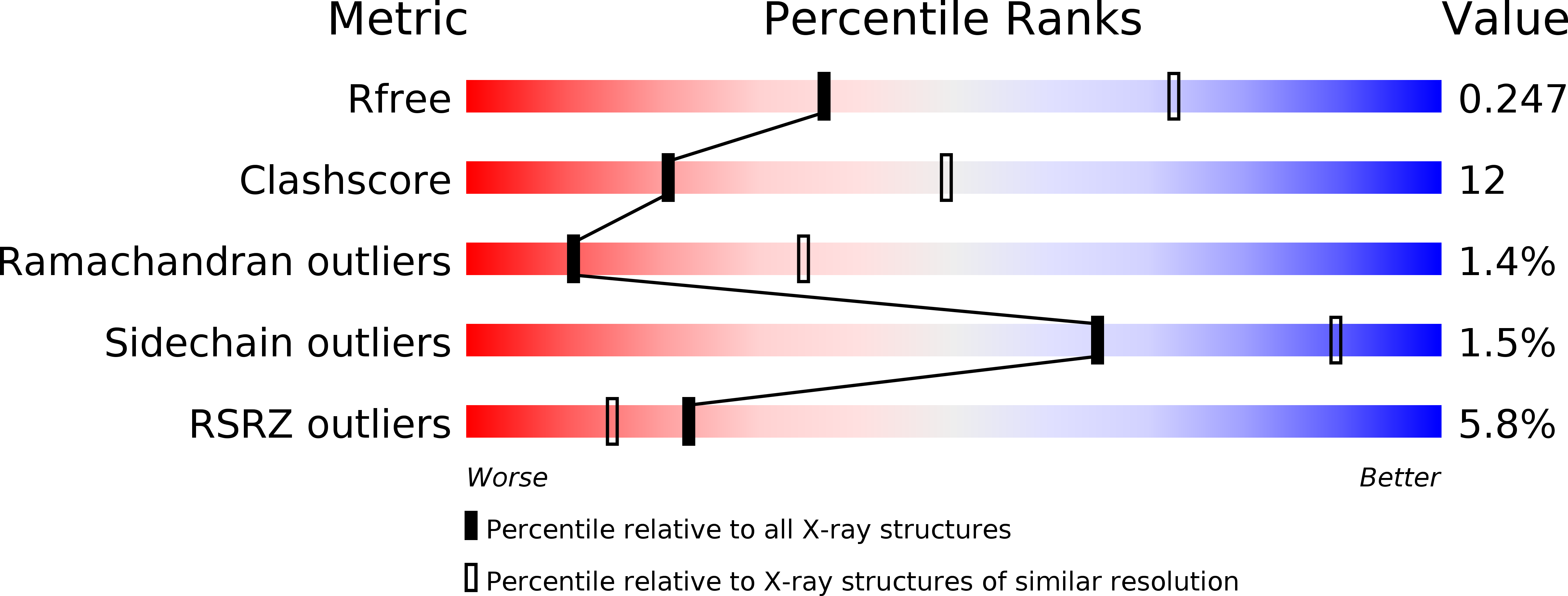
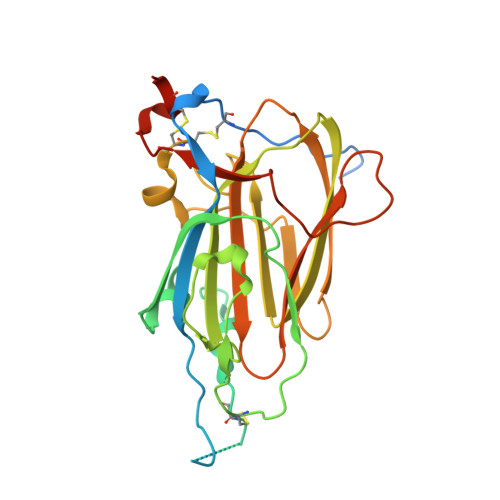
6Y98, 6Y9J - PubMed Abstract:
For host-cell interaction, the human fungal pathogen Candida glabrata harbors a large family of more than 20 cell wall-attached epithelial adhesins (Epas). Epa family members are lectins with binding pockets containing several conserved and variable structural hot spots, which were implicated in mediating functional diversity. In this study, we have performed an elaborate structure-based mutational analysis of numerous Epa paralogs to generally determine the role of diverse structural hot spots in conferring host cell binding and ligand binding specificity. Our study reveals that several conserved structural motifs contribute to efficient host cell binding. Moreover, our directed motif exchange experiments reveal that the variable loop CBL2 is key for programming ligand binding specificity, albeit with limited predictability. In contrast, we find that the variable loop L1 affects host cell binding without significantly influencing the specificity of ligand binding. Our data strongly suggest that variation of numerous structural hot spots in the ligand binding pocket of Epa proteins is a main driver of their functional diversification and evolution.
Organizational Affiliation:
Department of Genetics, Philipps-Universität, Marburg, Germany.