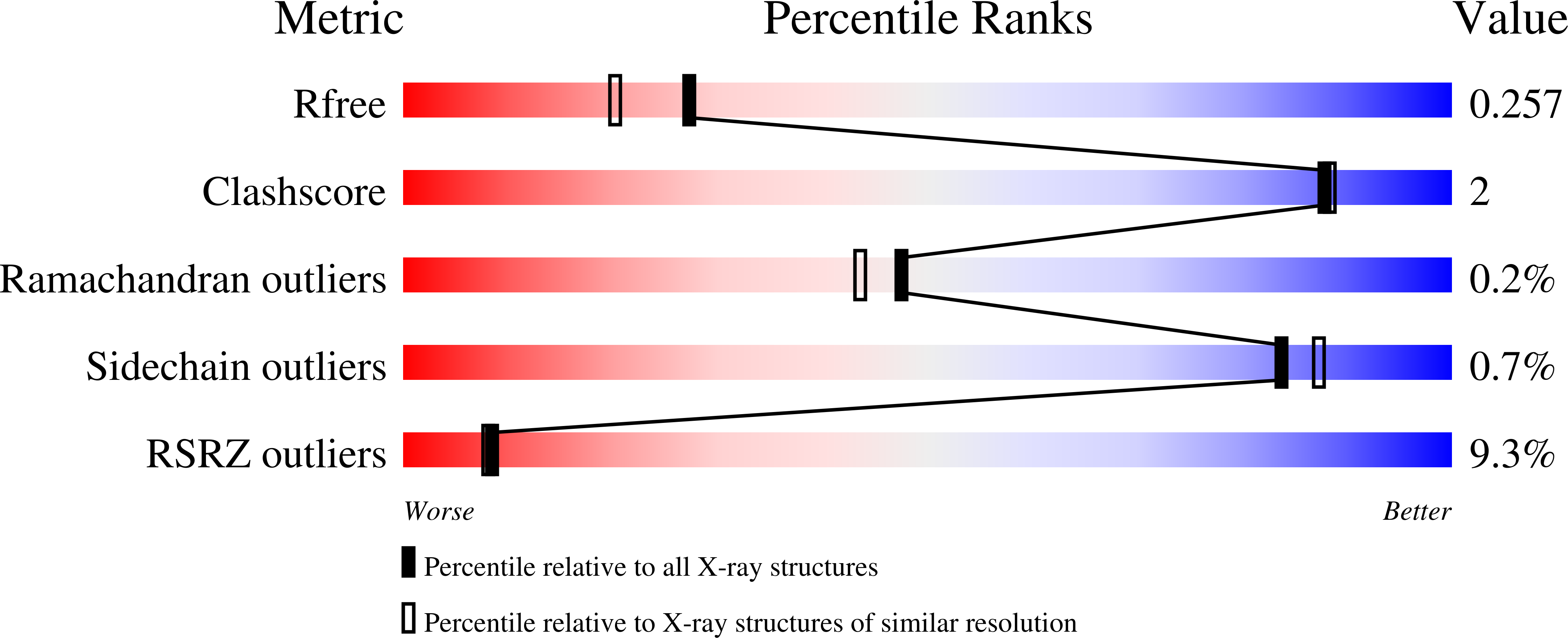
Molecular basis for ubiquitin ligase CRL2 FEM1C -mediated recognition of C-degron.
Yan, X., Wang, X., Li, Y., Zhou, M., Li, Y., Song, L., Mi, W., Min, J., Dong, C.(2021) Nat Chem Biol 17: 263-271
- PubMed: 33398170
- DOI: https://doi.org/10.1038/s41589-020-00703-4
- Primary Citation of Related Structures:
6XKC, 7JYA - PubMed Abstract:
Proteome integrity depends on the ubiquitin-proteasome system to degrade unwanted or abnormal proteins. In addition to the N-degrons, C-terminal residues of proteins can also serve as degradation signals (C-degrons) that are recognized by specific cullin-RING ubiquitin ligases (CRLs) for proteasomal degradation. FEM1C is a CRL2 substrate receptor that targets the C-terminal arginine degron (Arg/C-degron), but the molecular mechanism of substrate recognition remains largely elusive. Here, we present crystal structures of FEM1C in complex with Arg/C-degron and show that FEM1C utilizes a semi-open binding pocket to capture the C-terminal arginine and that the extreme C-terminal arginine is the major structural determinant in recognition by FEM1C. Together with biochemical and mutagenesis studies, we provide a framework for understanding molecular recognition of the Arg/C-degron by the FEM family of proteins.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, School of Basic Medical Sciences, Tianjin Medical University, Tianjin, China.