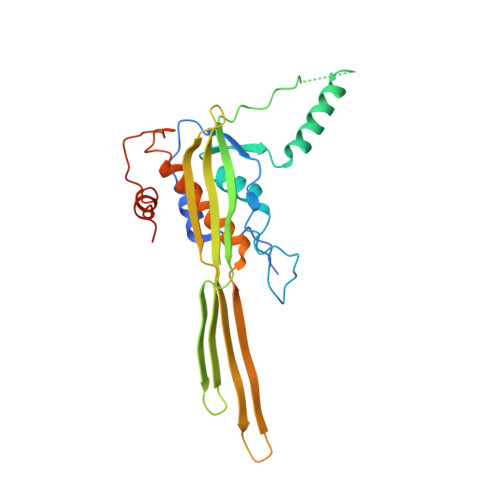
A dual-constriction biological nanopore resolves homonucleotide sequences with high fidelity.
Van der Verren, S.E., Van Gerven, N., Jonckheere, W., Hambley, R., Singh, P., Kilgour, J., Jordan, M., Wallace, E.J., Jayasinghe, L., Remaut, H.(2020) Nat Biotechnol 38: 1415-1420
- PubMed: 32632300
- DOI: https://doi.org/10.1038/s41587-020-0570-8
- Primary Citation of Related Structures:
6SI7 - PubMed Abstract:
Single-molecule long-read DNA sequencing with biological nanopores is fast and high-throughput but suffers reduced accuracy in homonucleotide stretches. We now combine the CsgG nanopore with the 35-residue N-terminal region of its extracellular interaction partner CsgF to produce a dual-constriction pore with improved signal and base-calling accuracy for homopolymer regions. The electron cryo-microscopy structure of CsgG in complex with full-length CsgF shows that the 33 N-terminal residues of CsgF bind inside the β-barrel of the pore, forming a defined second constriction. In complexes of CsgG bound to a 35-residue CsgF constriction peptide, the second constriction is separated from the primary constriction by ~25 Å. We find that both constrictions contribute to electrical signal modulation during single-stranded DNA translocation. DNA sequencing using a prototype CsgG-CsgF protein pore with two constrictions improved single-read accuracy by 25 to 70% in homopolymers up to 9 nucleotides long.
Organizational Affiliation:
Structural Biology Brussels, Vrije Universiteit Brussel, Brussels, Belgium.