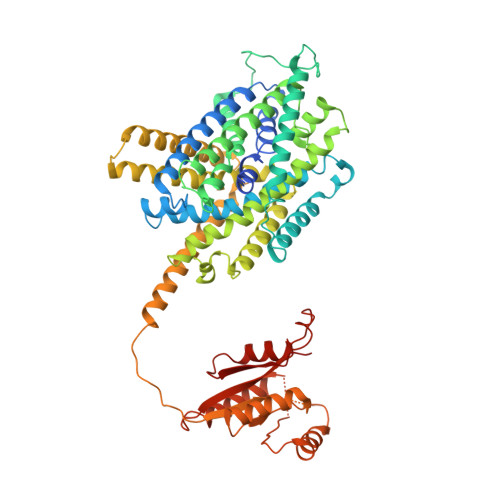
Structural basis of proton-coupled potassium transport in the KUP family.
Tascon, I., Sousa, J.S., Corey, R.A., Mills, D.J., Griwatz, D., Aumuller, N., Mikusevic, V., Stansfeld, P.J., Vonck, J., Hanelt, I.(2020) Nat Commun 11: 626-626
- PubMed: 32005818
- DOI: https://doi.org/10.1038/s41467-020-14441-7
- Primary Citation of Related Structures:
6S3K - PubMed Abstract:
Potassium homeostasis is vital for all organisms, but is challenging in single-celled organisms like bacteria and yeast and immobile organisms like plants that constantly need to adapt to changing external conditions. KUP transporters facilitate potassium uptake by the co-transport of protons. Here, we uncover the molecular basis for transport in this widely distributed family. We identify the potassium importer KimA from Bacillus subtilis as a member of the KUP family, demonstrate that it functions as a K + /H + symporter and report a 3.7 Å cryo-EM structure of the KimA homodimer in an inward-occluded, trans-inhibited conformation. By introducing point mutations, we identify key residues for potassium and proton binding, which are conserved among other KUP proteins.
Organizational Affiliation:
Institute of Biochemistry, Goethe University Frankfurt, Frankfurt am Main, Germany.