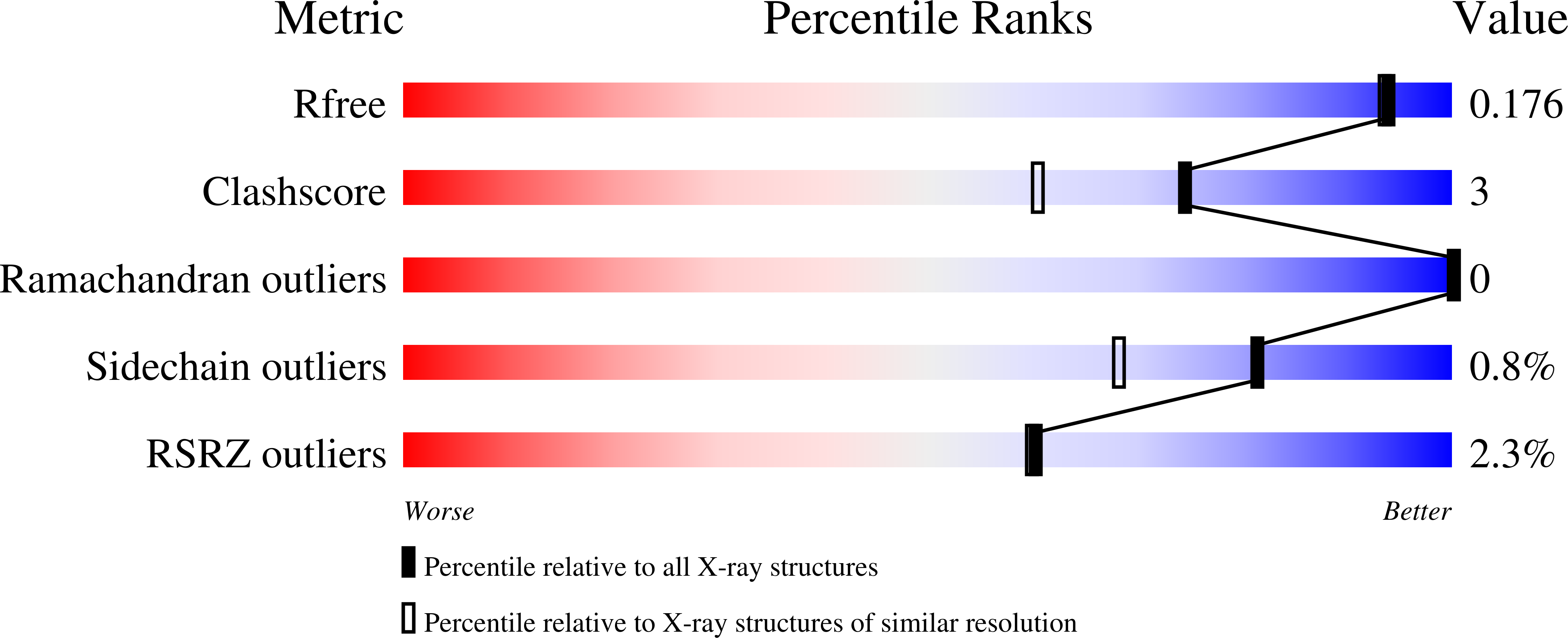
The dissociation mechanism of processive cellulases.
Vermaas, J.V., Kont, R., Beckham, G.T., Crowley, M.F., Gudmundsson, M., Sandgren, M., Stahlberg, J., Valjamae, P., Knott, B.C.(2019) Proc Natl Acad Sci U S A 116: 23061-23067
- PubMed: 31666327
- DOI: https://doi.org/10.1073/pnas.1913398116
- Primary Citation of Related Structures:
6RWF - PubMed Abstract:
Cellulase enzymes deconstruct recalcitrant cellulose into soluble sugars, making them a biocatalyst of biotechnological interest for use in the nascent lignocellulosic bioeconomy. Cellobiohydrolases (CBHs) are cellulases capable of liberating many sugar molecules in a processive manner without dissociating from the substrate. Within the complete processive cycle of CBHs, dissociation from the cellulose substrate is rate limiting, but the molecular mechanism of this step is unknown. Here, we present a direct comparison of potential molecular mechanisms for dissociation via Hamiltonian replica exchange molecular dynamics of the model fungal CBH, Trichoderma reesei Cel7A. Computational rate estimates indicate that stepwise cellulose dethreading from the binding tunnel is 4 orders of magnitude faster than a clamshell mechanism, in which the substrate-enclosing loops open and release the substrate without reversing. We also present the crystal structure of a disulfide variant that covalently links substrate-enclosing loops on either side of the substrate-binding tunnel, which constitutes a CBH that can only dissociate via stepwise dethreading. Biochemical measurements indicate that this variant has a dissociation rate constant essentially equivalent to the wild type, implying that dethreading is likely the predominant mechanism for dissociation.
Organizational Affiliation:
Biosciences Center, National Renewable Energy Laboratory, Golden, CO 80401.