Evidence of Destabilization of the Human Thymidylate Synthase (hTS) Dimeric Structure Induced by the Interface Mutation Q62R.
Pozzi, C., Lopresti, L., Santucci, M., Costi, M.P., Mangani, S.(2019) Biomolecules 9
- PubMed: 30987202
- DOI: https://doi.org/10.3390/biom9040134
- Primary Citation of Related Structures:
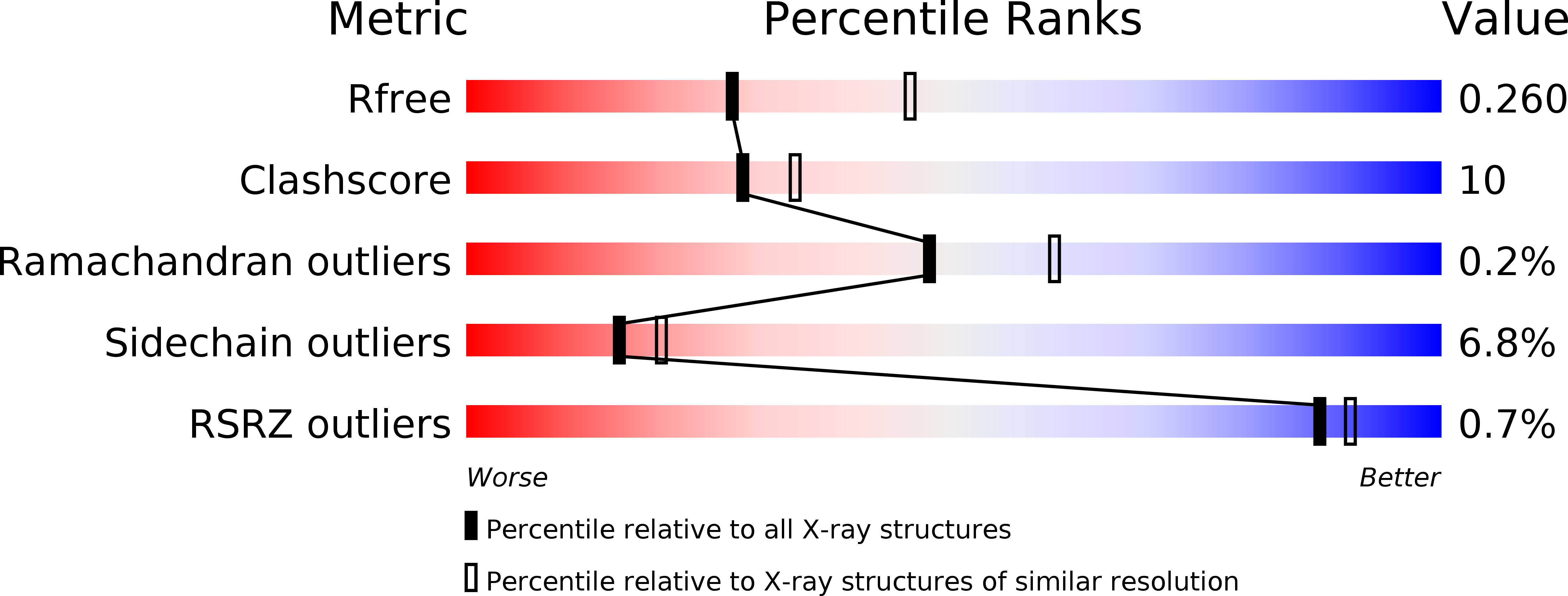
6R2E - PubMed Abstract:
In human cells, thymidylate synthase (TS) provides the only source of 2'-deoxythymidyne-5'-monophosphate (dTMP), which is required for DNA biosynthesis. Because of its pivotal role, human TS (hTS) represents a validated target for anticancer chemotherapy. Nonetheless, the efficacy of drugs blocking the hTS active site has limitations due to the onset of resistance in cancer cells, requiring the identification of new strategies to effectively inhibit this enzyme. Human TS works as an obligate homodimer, making the inter-subunit interface an attractive targetable area. Here, we report the design and investigation of a new hTS variant, in which Gln62, located at the dimer interface, has been replaced by arginine in order to destabilize the enzyme quaternary assembly. The hTS Q62R variant has been characterized though kinetic assay, thermal denaturation analysis and X-ray crystallography. Our results provide evidence that hTS Q62R has a reduced melting temperature. The effective destabilization of the TS quaternary structure is also confirmed by structural analysis, showing that the introduced mutation induces a slight aperture of the hTS dimer. The generation of hTS variants having a more accessible interface area can facilitate the screening of interface-targeting molecules, providing key information for the rational design of innovative hTS interface inhibitors.
Organizational Affiliation:
Department of Biotechnology, Chemistry and Pharmacy-Department of Excellence 2018-2020, University of Siena, 53100 Siena, Italy. pozzi4@unisi.it.