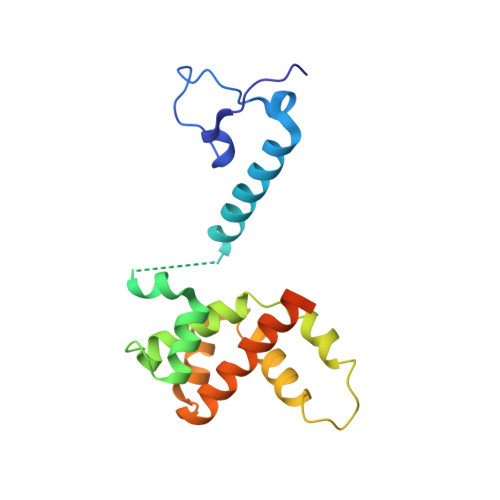
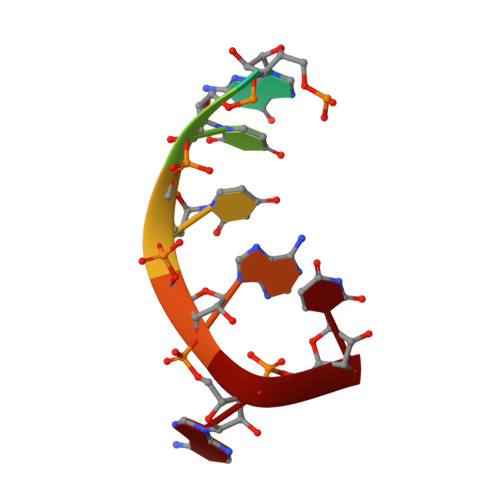
Structure of the Human Respiratory Syncytial Virus M2-1 Protein in Complex with a Short Positive-Sense Gene-End RNA.
Gao, Y., Cao, D., Pawnikar, S., John, K.P., Ahn, H.M., Hill, S., Ha, J.M., Parikh, P., Ogilvie, C., Swain, A., Yang, A., Bell, A., Salazar, A., Miao, Y., Liang, B.(2020) Structure 28: 979
- PubMed: 32697936
- DOI: https://doi.org/10.1016/j.str.2020.07.001
- Primary Citation of Related Structures:
6PZQ - PubMed Abstract:
The M2-1 protein of human respiratory syncytial virus (HRSV) is a transcription anti-terminator that regulates the processivity of the HRSV RNA-dependent RNA polymerase (RdRP). Here, we report a crystal structure of HRSV M2-1 bound to a short positive-sense gene-end RNA (SH7) at 2.7 Å resolution. We identified multiple critical residues of M2-1 involved in RNA interaction and examined their roles using mutagenesis and MicroScale Thermophoresis (MST) assay. We found that hydrophobic residue Phe23 is indispensable for M2-1 to recognize the base of RNA. We also captured spontaneous binding of RNA (SH7) to M2-1 in all-atom simulations using a robust Gaussian accelerated molecular dynamics (GaMD) method. Both experiments and simulations revealed that the interactions of RNA with two separate domains of M2-1, the zinc-binding domain (ZBD) and the core domain (CD), are independent of each other. Collectively, our results provided a structural basis for RNA recognition by HRSV M2-1.
Organizational Affiliation:
Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322 USA.