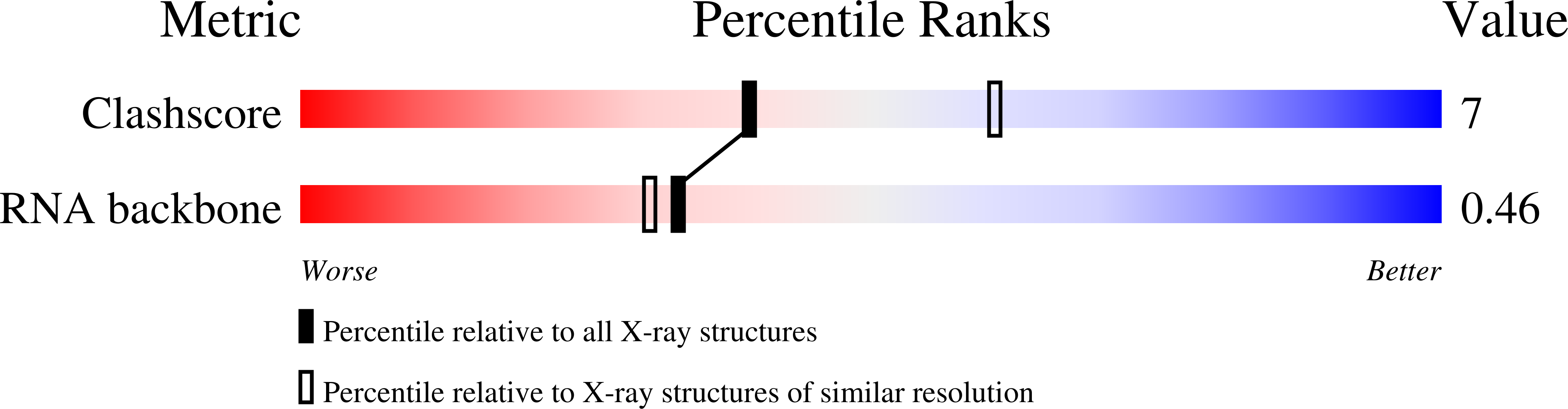Ribosomal Protein L11 Selectively Stabilizes a Tertiary Structure of the GTPase Center rRNA Domain.
Welty, R., Rau, M., Pabit, S., Dunstan, M.S., Conn, G.L., Pollack, L., Hall, K.B.(2020) J Mol Biol 432: 991-1007
- PubMed: 31874150
- DOI: https://doi.org/10.1016/j.jmb.2019.12.010
- Primary Citation of Related Structures:
6PRV - PubMed Abstract:
The GTPase Center (GAC) RNA domain in bacterial 23S rRNA is directly bound by ribosomal protein L11, and this complex is essential to ribosome function. Previous cocrystal structures of the 58-nucleotide GAC RNA bound to L11 revealed the intricate tertiary fold of the RNA domain, with one monovalent and several divalent ions located in specific sites within the structure. Here, we report a new crystal structure of the free GAC that is essentially identical to the L11-bound structure, which retains many common sites of divalent ion occupation. This new structure demonstrates that RNA alone folds into its tertiary structure with bound divalent ions. In solution, we find that this tertiary structure is not static, but rather is best described as an ensemble of states. While L11 protein cannot bind to the GAC until the RNA has adopted its tertiary structure, new experimental data show that L11 binds to Mg 2+ -dependent folded states, which we suggest lie along the folding pathway of the RNA. We propose that L11 stabilizes a specific GAC RNA tertiary state, corresponding to the crystal structure, and that this structure reflects the functionally critical conformation of the rRNA domain in the fully assembled ribosome.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, 660 S Euclid Ave, St Louis, MO, 63110, USA; Department of Chemistry, University of Michigan, Ann Arbor, MI, 48109, USA.