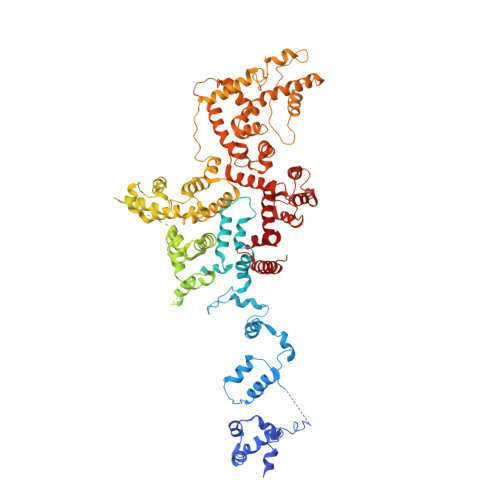
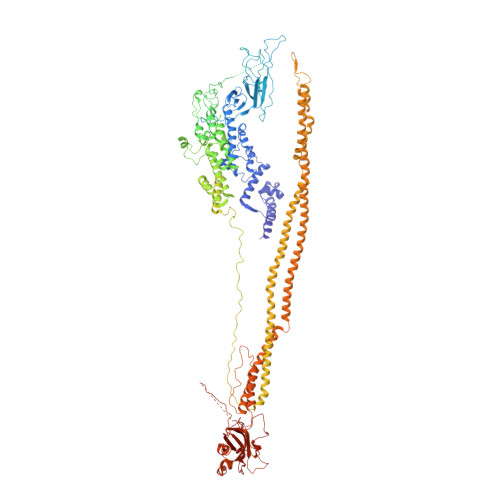
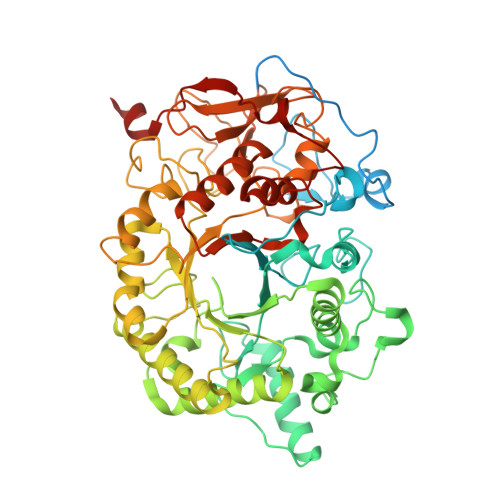
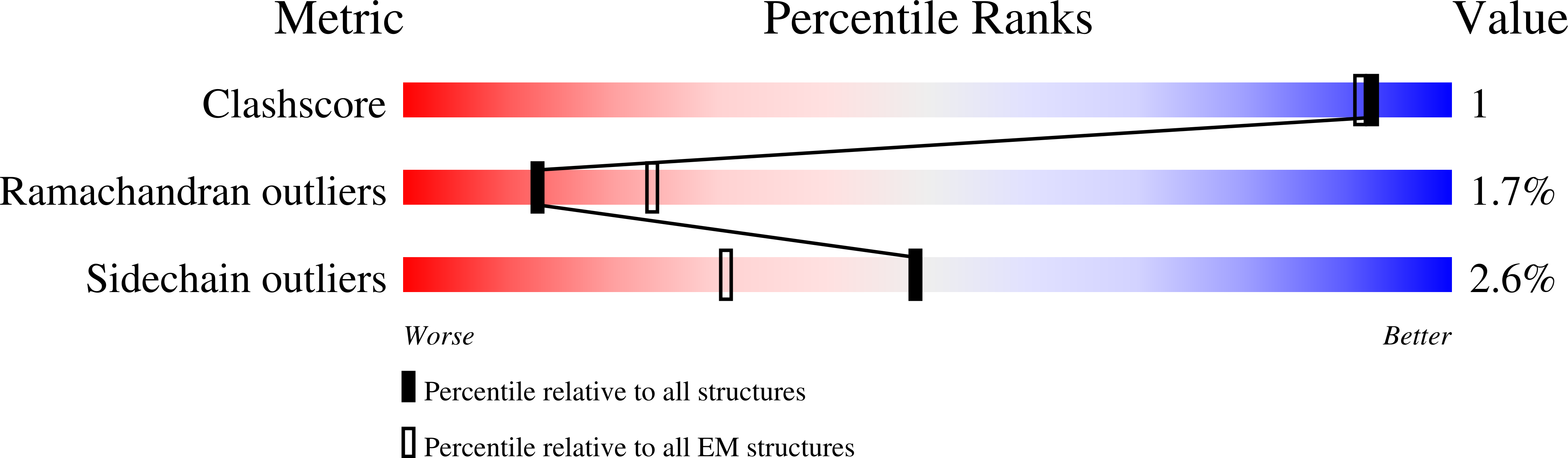Cryo-EM structures of the pore-forming A subunit from the Yersinia entomophaga ABC toxin.
Piper, S.J., Brillault, L., Rothnagel, R., Croll, T.I., Box, J.K., Chassagnon, I., Scherer, S., Goldie, K.N., Jones, S.A., Schepers, F., Hartley-Tassell, L., Ve, T., Busby, J.N., Dalziel, J.E., Lott, J.S., Hankamer, B., Stahlberg, H., Hurst, M.R.H., Landsberg, M.J.(2019) Nat Commun 10: 1952-1952
- PubMed: 31028251
- DOI: https://doi.org/10.1038/s41467-019-09890-8
- Primary Citation of Related Structures:
6OGD - PubMed Abstract:
ABC toxins are pore-forming virulence factors produced by pathogenic bacteria. YenTcA is the pore-forming and membrane binding A subunit of the ABC toxin YenTc, produced by the insect pathogen Yersinia entomophaga. Here we present cryo-EM structures of YenTcA, purified from the native source. The soluble pre-pore structure, determined at an average resolution of 4.4 Å, reveals a pentameric assembly that in contrast to other characterised ABC toxins is formed by two TcA-like proteins (YenA1 and YenA2) and decorated by two endochitinases (Chi1 and Chi2). We also identify conformational changes that accompany membrane pore formation by visualising YenTcA inserted into liposomes. A clear outward rotation of the Chi1 subunits allows for access of the protruding translocation pore to the membrane. Our results highlight structural and functional diversity within the ABC toxin subfamily, explaining how different ABC toxins are capable of recognising diverse hosts.
Organizational Affiliation:
School of Chemistry and Molecular Biosciences, The University of Queensland, St Lucia Queensland, 4072, Australia.