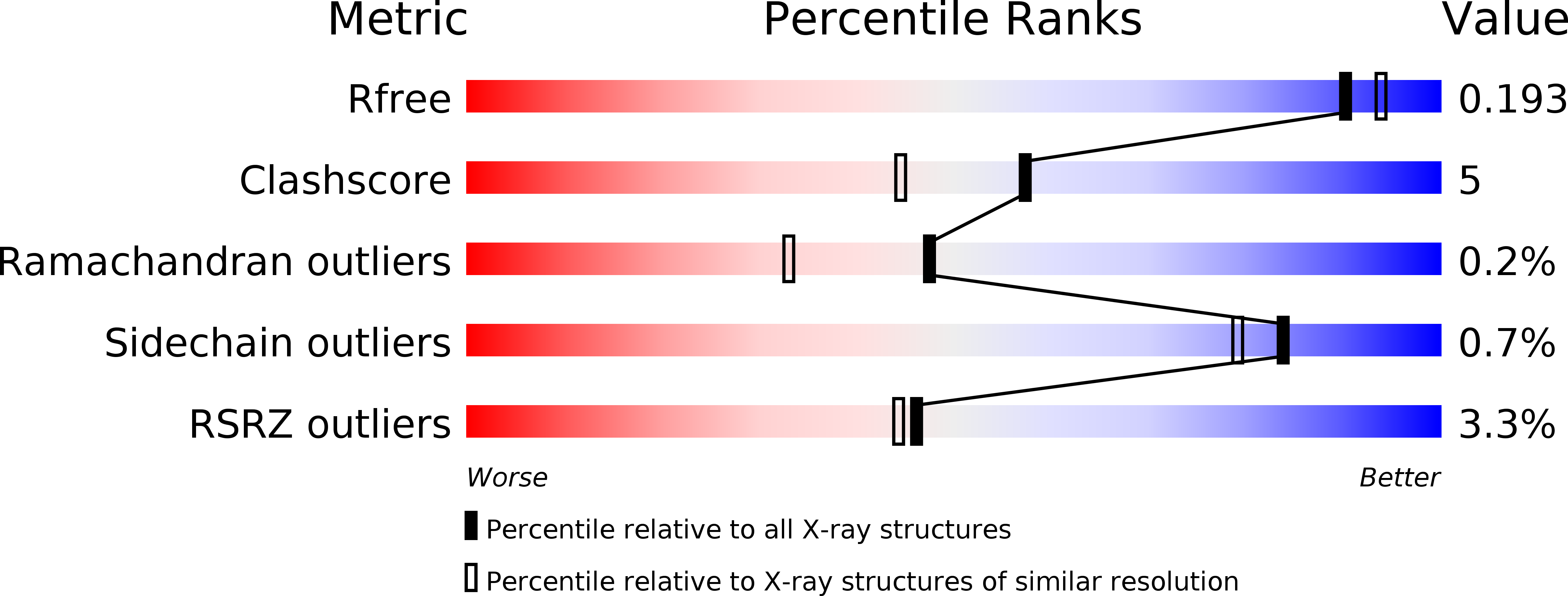
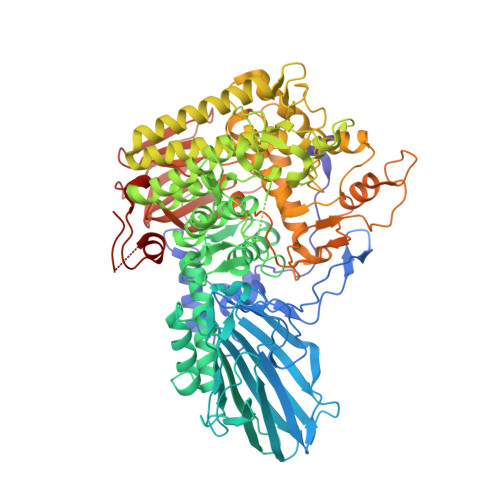
Crystal structure of beta-L-arabinobiosidase belonging to glycoside hydrolase family 121.
Saito, K., Viborg, A.H., Sakamoto, S., Arakawa, T., Yamada, C., Fujita, K., Fushinobu, S.(2020) PLoS One 15: e0231513-e0231513
- PubMed: 32479540
- DOI: https://doi.org/10.1371/journal.pone.0231513
- Primary Citation of Related Structures:
6M5A - PubMed Abstract:
Enzymes acting on α-L-arabinofuranosides have been extensively studied; however, the structures and functions of β-L-arabinofuranosidases are not fully understood. Three enzymes and an ABC transporter in a gene cluster of Bifidobacterium longum JCM 1217 constitute a degradation and import system of β-L-arabinooligosaccharides on plant hydroxyproline-rich glycoproteins. An extracellular β-L-arabinobiosidase (HypBA2) belonging to the glycoside hydrolase (GH) family 121 plays a key role in the degradation pathway by releasing β-1,2-linked arabinofuranose disaccharide (β-Ara2) for the specific sugar importer. Here, we present the crystal structure of the catalytic region of HypBA2 as the first three-dimensional structure of GH121 at 1.85 Å resolution. The HypBA2 structure consists of a central catalytic (α/α)6 barrel domain and two flanking (N- and C-terminal) β-sandwich domains. A pocket in the catalytic domain appears to be suitable for accommodating the β-Ara2 disaccharide. Three acidic residues Glu383, Asp515, and Glu713, located in this pocket, are completely conserved among all members of GH121; site-directed mutagenesis analysis showed that they are essential for catalytic activity. The active site of HypBA2 was compared with those of structural homologs in other GH families: GH63 α-glycosidase, GH94 chitobiose phosphorylase, GH142 β-L-arabinofuranosidase, GH78 α-L-rhamnosidase, and GH37 α,α-trehalase. Based on these analyses, we concluded that the three conserved residues are essential for catalysis and substrate binding. β-L-Arabinobiosidase genes in GH121 are mainly found in the genomes of bifidobacteria and Xanthomonas species, suggesting that the cleavage and specific import system for the β-Ara2 disaccharide on plant hydroxyproline-rich glycoproteins are shared in animal gut symbionts and plant pathogens.
Organizational Affiliation:
Department of Biotechnology, The University of Tokyo, Tokyo, Japan.