Regio- and stereoselective hydroxylation of testosterone by a novel cytochrome P450 154C2 from Streptomyces avermitilis.
Wang, Q., Ma, B., Fushinobu, S., Zhang, C., Xu, L.H.(2020) Biochem Biophys Res Commun 522: 355-361
- PubMed: 31767148
- DOI: https://doi.org/10.1016/j.bbrc.2019.11.091
- Primary Citation of Related Structures:
6L69 - PubMed Abstract:
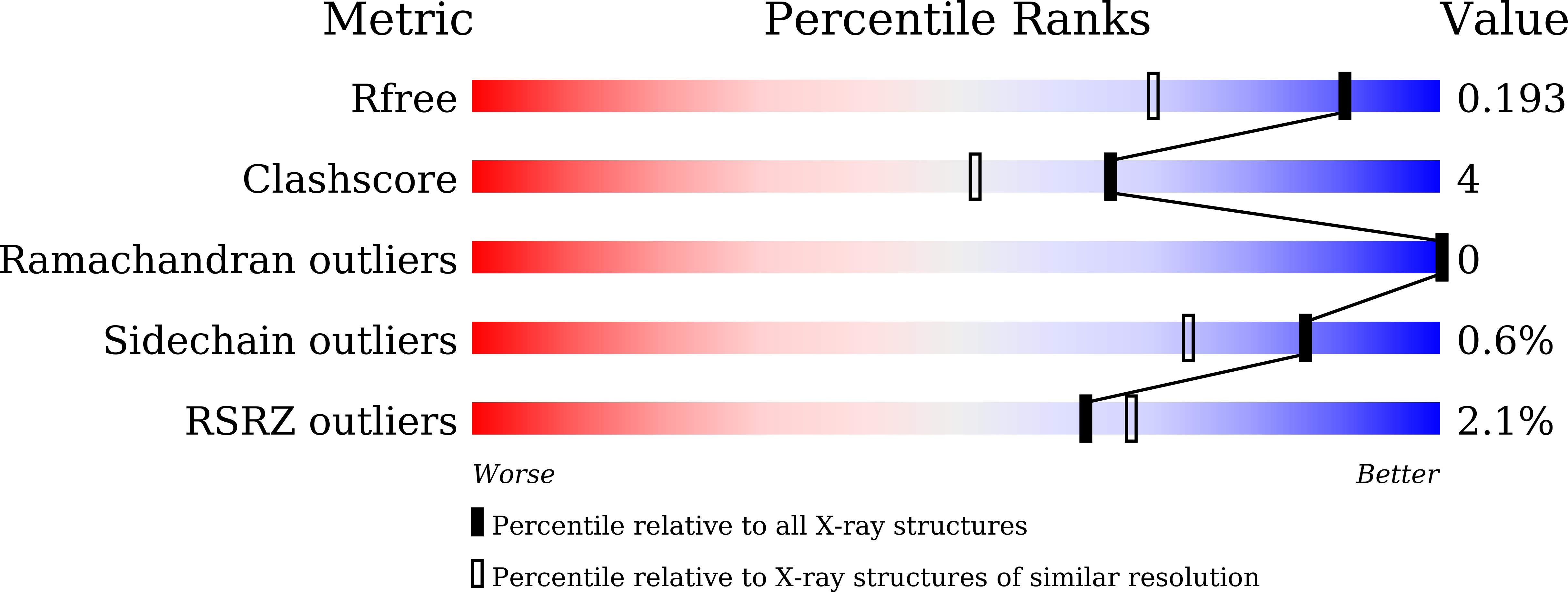
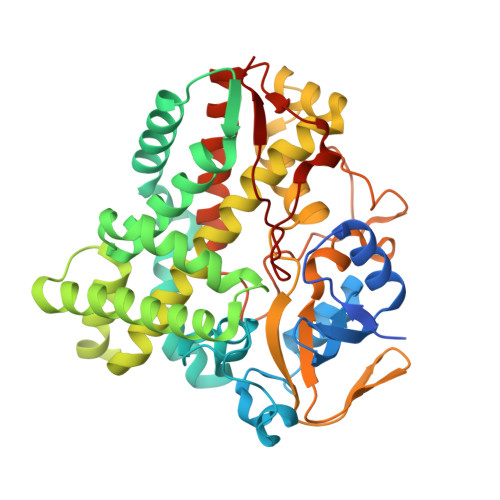
Cytochrome P450 enzymes (P450 or CYP) are some of the most versatile biocatalysts, and offer advantages for oxidizing unreactive C-H bonds in mild conditions. In this study, we identified a novel cytochrome P450 154C2 from Streptomyces avermitilis and characterized its function in 2α-hydroxylation of testosterone with regio- and stereoselectivity. To investigate the efficiency of electron transfer, we conducted biotransformation using two different P450 redox partners-RhFRED (RhF reductase domain) from Rhodococcus sp. and Pdx (putidaredoxin)/Pdr (putidaredoxin reductase) from Pseudomonas putida and revealed that RhFRED was more effective than Pdx/Pdr, especially in vivo. The K m and k cat values for testosterone were estimated to be 0.16 ± 0.05 mM and 0.13 ± 0.02 min -1 , and k cat /K m was 0.81 min -1 mM -1 . We also determined the crystal structure of the substrate-free form of CYP154C2 at 1.5 Å resolution. The structure has a closed conformation, and the substrate binding pocket is narrow, which can explain the strict substrate specificity of the enzyme.
Organizational Affiliation:
Ocean College, Zhejiang University, Dinghai, Zhoushan, Zhejiang, 316021, China.