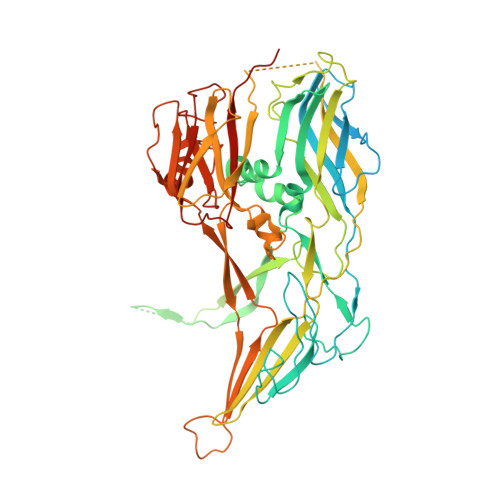
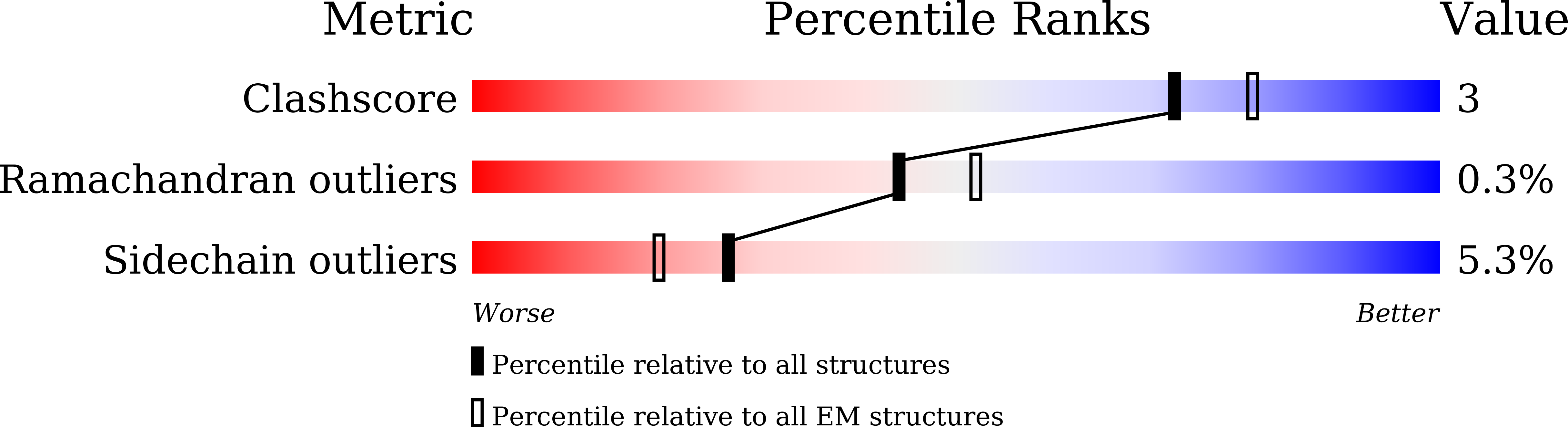Structure of the African swine fever virus major capsid protein p72.
Liu, Q., Ma, B., Qian, N., Zhang, F., Tan, X., Lei, J., Xiang, Y.(2019) Cell Res 29: 953-955
- PubMed: 31530894
- DOI: https://doi.org/10.1038/s41422-019-0232-x
- Primary Citation of Related Structures:
6KU9
Organizational Affiliation:
Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, Center for Infectious Disease Research, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing, 100084, China.