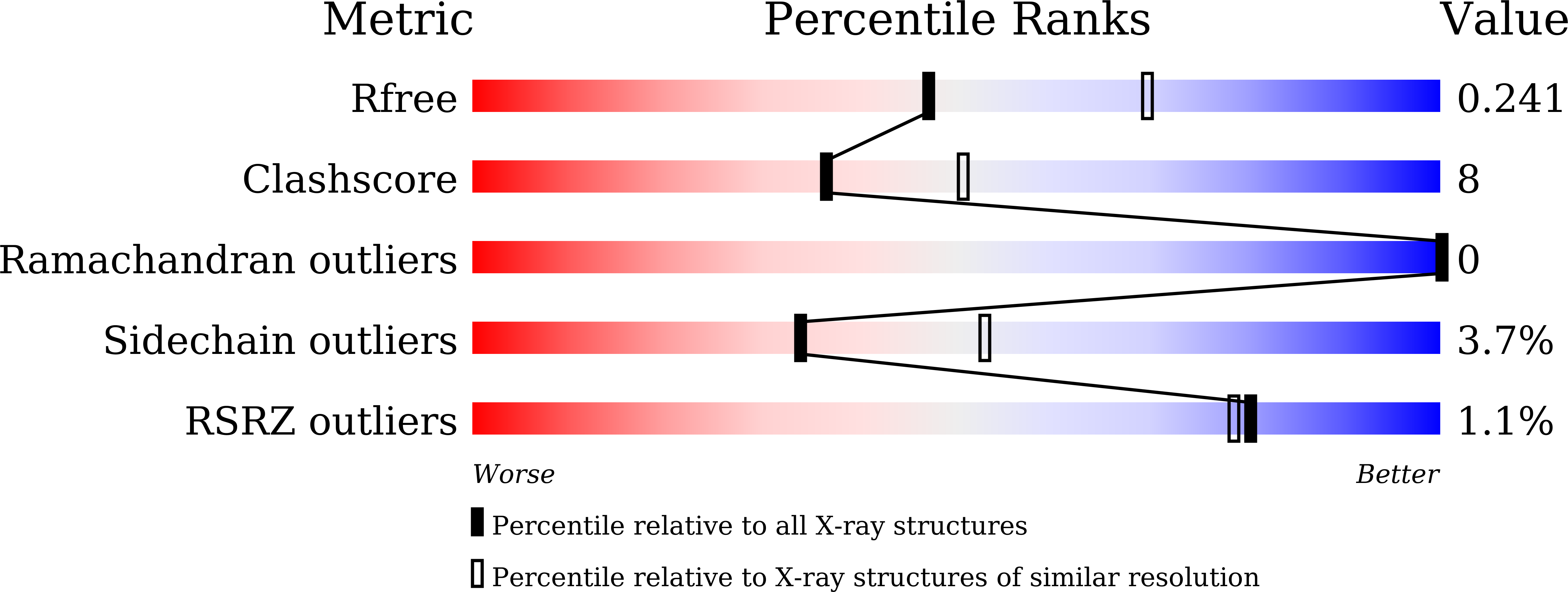
The Structural Basis for Specific Recognition of H3K14 Acetylation by Sth1 in the RSC Chromatin Remodeling Complex.
Chen, G., Li, W., Yan, F., Wang, D., Chen, Y.(2020) Structure 28: 111
- PubMed: 31711754
- DOI: https://doi.org/10.1016/j.str.2019.10.015
- Primary Citation of Related Structures:
6KMB, 6KMJ - PubMed Abstract:
The Saccharomyces cerevisiae RSC (Remodel the Structure of Chromatin) complex is a chromatin-remodeling complex and plays essential roles in transcription regulation and DNA repair. The acetylation of H3 Lysine14 (H3K14Ac) enhances the RSC retention on nucleosomes and increases the remodeling activity of RSC. However, which RSC component recognizes H3K14Ac remains unclear. Here, we discovered that the bromodomain of the catalytic subunit Sth1 (Sth1 BD ) possessed the strongest affinity to H3K14Ac among all RSC bromodomains. The Sth1 BD specifically recognized the K(Ac)ΦΦR motif (Φ stands for any hydrophobic amino acid), including H3K14Ac and H4K20Ac. We determined the crystal structures of Sth1 BD at 2.40 Å resolution and Sth1 BD -H3K14Ac complex at 1.40 Å resolution. The extensive interfaces between Sth1 BD and H3 6-21 facilitate the specific and robust binding of Sth1 BD to H3K14Ac. Our studies provide insights into how the RSC complex recognizes H3K14Ac to orchestrate the crosstalk between histone acetylation and chromatin remodeling.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, National Center for Protein Science Shanghai, Shanghai Science Research Center, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, University of Chinese Academy of Sciences, 333 Haike Road, Shanghai 201210, China.