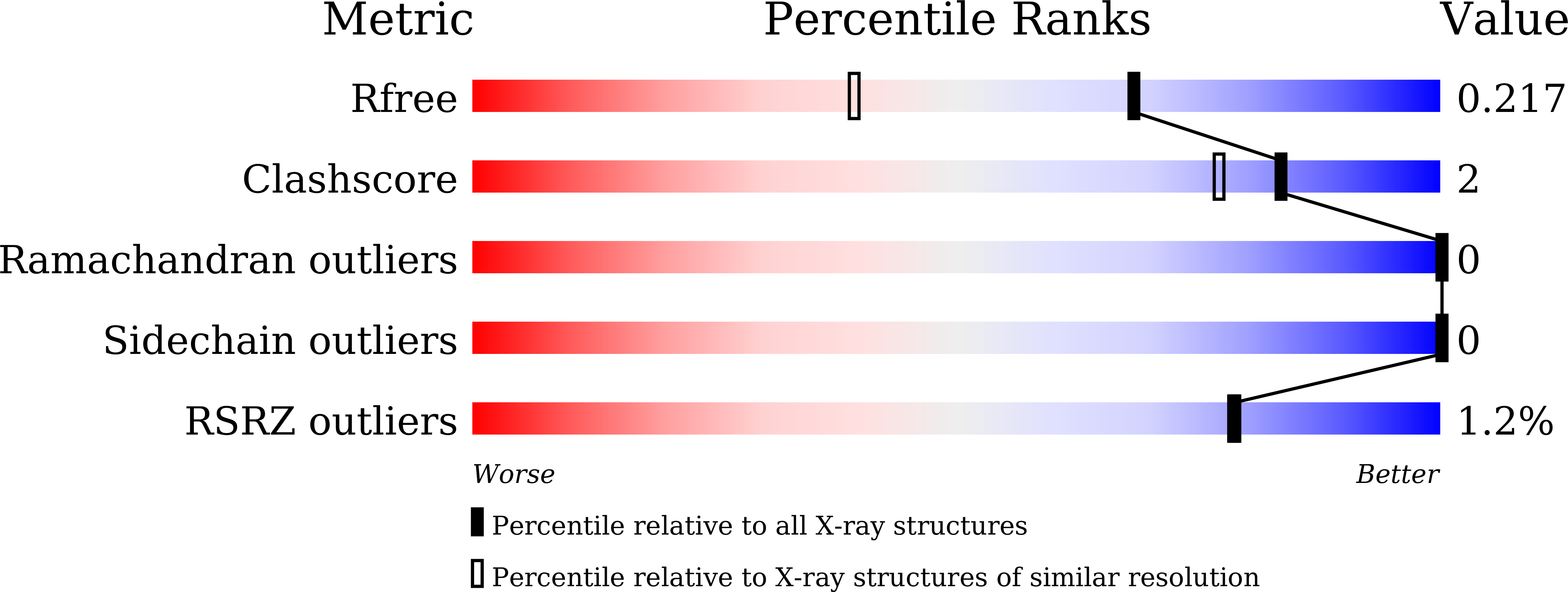
Grafting a short chameleon sequence from alpha B crystallin into a beta-sheet scaffold protein.
Hori, Y., Fujiwara, H., Fujiwara, W., Makabe, K.(2019) Proteins 87: 416-424
- PubMed: 30684364
- DOI: https://doi.org/10.1002/prot.25663
- Primary Citation of Related Structures:
6J47, 6J48, 6J49 - PubMed Abstract:
Many protein and peptide sequences are self-assembled into β-sheet-rich fibrous structures called amyloids. Their atomic details provide insights into fundamental knowledge related to amyloid diseases. To study the detailed structure of the amyloid, we have developed a model system that mimics the self-assembling process of the amyloid within a water-soluble protein, termed peptide self-assembly mimic (PSAM). PSAM enables capturing of a peptide sequence within a water-soluble protein, thus making structural and energetics-related studies possible. In this work, we extend our PSAM approach to a naturally occurring chameleon sequence from αB crystallin. We chose "Val-Leu-Gly-Asp-Val (VLGDV)", a five amino-acid sequence, which forms a β-turn in the native structure and a β-barrel in the amyloid oligomer cylindrin, as a grafting sequence to the PSAM scaffold. The crystal structure revealed that the sequence grafting induced β-sheet bending at the grafted site. We further investigated the role of the central glycine residue and found that its role in the β-sheet bending is dependent on the neighboring residues. The ability of PSAM to observe the structural alterations induced by the grafted sequence provides an opportunity to evaluate the structural impact of a sequence from the peptide self-assembly.
Organizational Affiliation:
Graduate School of Science and Engineering, Yamagata University, Yonezawa, Yamagata, Japan.